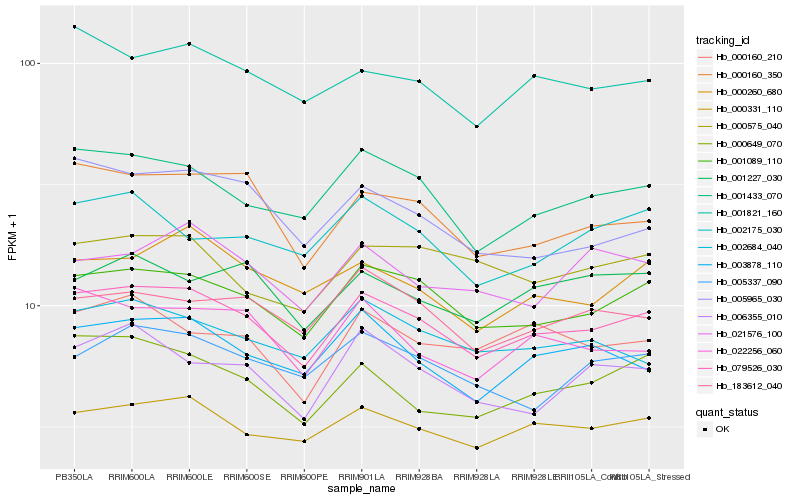
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079526_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 2 |
Hb_001089_110 |
0.0383811324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000331_110 |
0.0499632138 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 4 |
Hb_001433_070 |
0.0515744364 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 5 |
Hb_003878_110 |
0.0632805904 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 6 |
Hb_001821_160 |
0.0634148181 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 7 |
Hb_000160_350 |
0.0638407579 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 8 |
Hb_183612_040 |
0.0660309737 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 9 |
Hb_000260_680 |
0.0664006428 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 10 |
Hb_002684_040 |
0.0675366458 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 11 |
Hb_022256_060 |
0.0677168711 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 12 |
Hb_000649_070 |
0.0686181332 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 13 |
Hb_005337_090 |
0.0692819347 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 14 |
Hb_000160_210 |
0.0704888182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006355_010 |
0.0705611121 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 16 |
Hb_001227_030 |
0.071032448 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000575_040 |
0.071944442 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 18 |
Hb_002175_030 |
0.0721982623 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 19 |
Hb_021576_100 |
0.073472509 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 20 |
Hb_005965_030 |
0.0750455426 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |