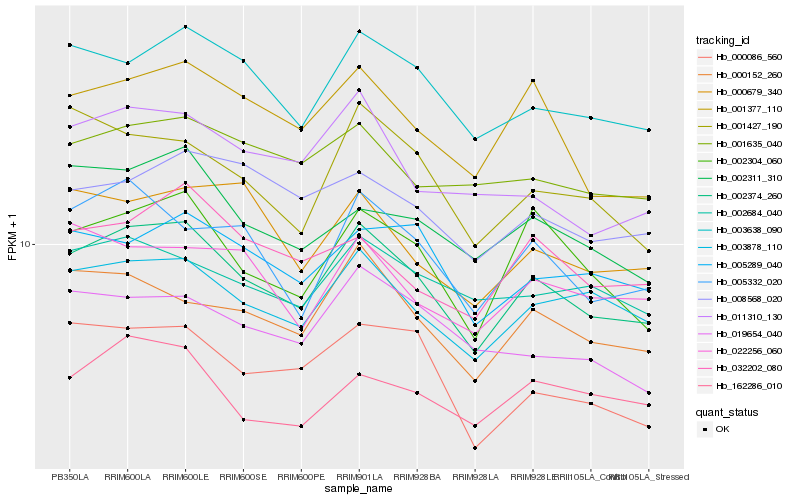
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_260 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 2 |
Hb_003878_110 |
0.0641994223 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 3 |
Hb_003638_090 |
0.0652896217 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 4 |
Hb_000152_260 |
0.0700835782 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 5 |
Hb_002304_060 |
0.0701465233 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 6 |
Hb_001377_110 |
0.0712572246 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 7 |
Hb_005332_020 |
0.0739788334 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 8 |
Hb_002311_310 |
0.0794792749 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 9 |
Hb_001427_190 |
0.0799873424 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 10 |
Hb_011310_130 |
0.0808580195 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_005289_040 |
0.080922125 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 12 |
Hb_032202_080 |
0.0827593384 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002684_040 |
0.083788202 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001635_040 |
0.0845103872 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_000086_560 |
0.0846938789 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_019654_040 |
0.0848043852 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 17 |
Hb_162286_010 |
0.0852362771 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 18 |
Hb_000679_340 |
0.0852645922 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008568_020 |
0.0857237189 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 20 |
Hb_022256_060 |
0.0864678435 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |