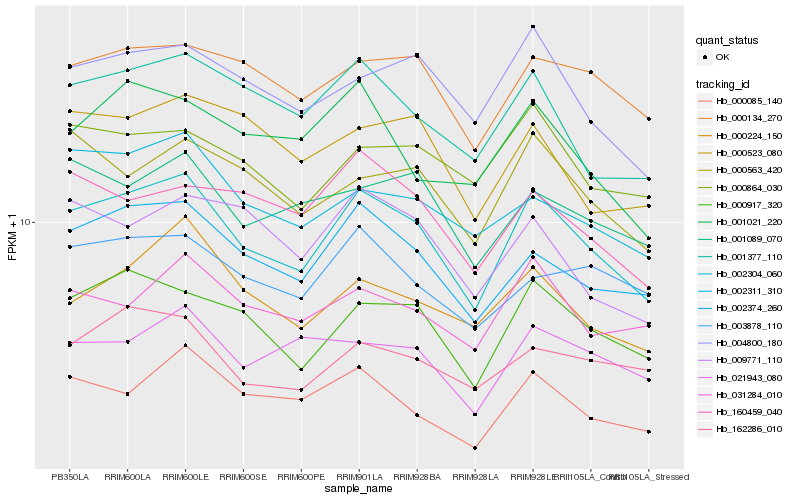
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_060 |
0.0 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 2 |
Hb_002374_260 |
0.0701465233 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 3 |
Hb_000917_320 |
0.0711249552 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001377_110 |
0.0822768466 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 5 |
Hb_004800_180 |
0.0856431224 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 6 |
Hb_000224_150 |
0.0877375329 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000085_140 |
0.0879338927 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 8 |
Hb_000134_270 |
0.089803917 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 9 |
Hb_000563_420 |
0.0908585765 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 10 |
Hb_031284_010 |
0.0909414305 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 11 |
Hb_002311_310 |
0.0927219174 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 12 |
Hb_001089_070 |
0.0932956464 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 13 |
Hb_162286_010 |
0.0936062816 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 14 |
Hb_000864_030 |
0.0949220449 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003878_110 |
0.0952416685 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 16 |
Hb_021943_080 |
0.0953069277 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001021_220 |
0.0967806943 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 18 |
Hb_160459_040 |
0.0968121618 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 19 |
Hb_000523_080 |
0.0975750376 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 20 |
Hb_009771_110 |
0.0976830847 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |