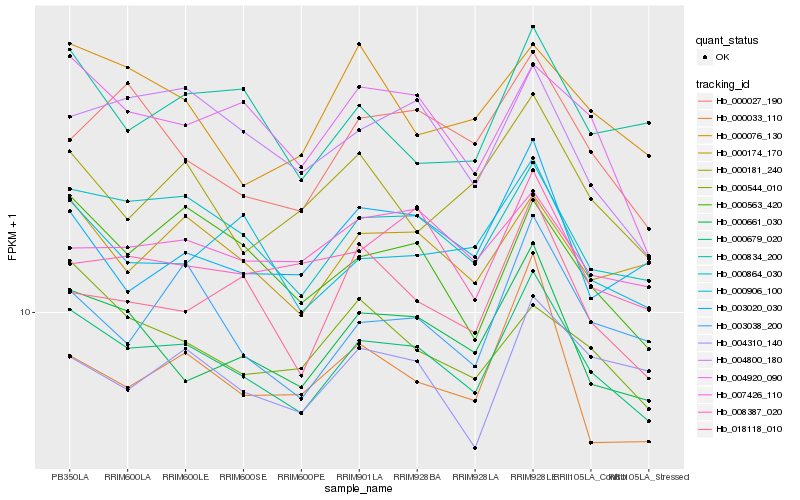
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_020 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 2 |
Hb_000864_030 |
0.0498441104 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000661_030 |
0.0671613718 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 4 |
Hb_000174_170 |
0.072429687 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000563_420 |
0.0762219756 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 6 |
Hb_004920_090 |
0.0762982171 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000906_100 |
0.0785744763 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 8 |
Hb_003038_200 |
0.0807990606 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 9 |
Hb_018118_010 |
0.0816452408 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 10 |
Hb_004800_180 |
0.0819346598 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 11 |
Hb_000544_010 |
0.0856179534 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000027_190 |
0.0870214341 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000834_200 |
0.087206887 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 14 |
Hb_008387_020 |
0.0877940729 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 15 |
Hb_007426_110 |
0.0878201608 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000181_240 |
0.0883049009 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 17 |
Hb_003020_030 |
0.0885073539 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_000033_110 |
0.0885701639 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 19 |
Hb_004310_140 |
0.0886841212 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_000076_130 |
0.0893679175 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |