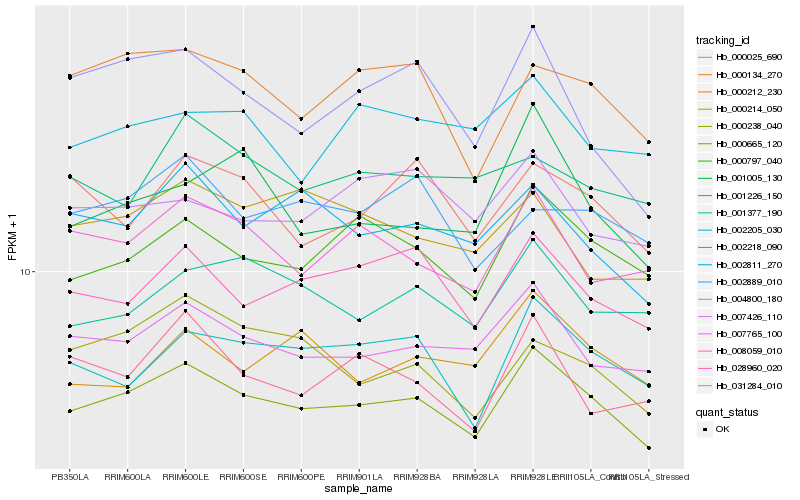
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_120 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 2 |
Hb_007765_100 |
0.0543279833 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 3 |
Hb_000797_040 |
0.05861656 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 4 |
Hb_028960_020 |
0.0642491232 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002205_030 |
0.0656827601 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 6 |
Hb_008059_010 |
0.0676312561 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002811_270 |
0.0708871046 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 8 |
Hb_001377_190 |
0.0711387747 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000134_270 |
0.0728852458 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 10 |
Hb_000025_690 |
0.0772437148 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 11 |
Hb_031284_010 |
0.0775304331 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 12 |
Hb_000212_230 |
0.0777804163 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 13 |
Hb_002218_090 |
0.0784293936 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 14 |
Hb_007426_110 |
0.0784417975 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_000238_040 |
0.0796063116 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 16 |
Hb_001005_130 |
0.0798943909 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 17 |
Hb_004800_180 |
0.0799921583 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 18 |
Hb_002889_010 |
0.0804865337 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 19 |
Hb_000214_050 |
0.0817403063 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001226_150 |
0.0818673658 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |