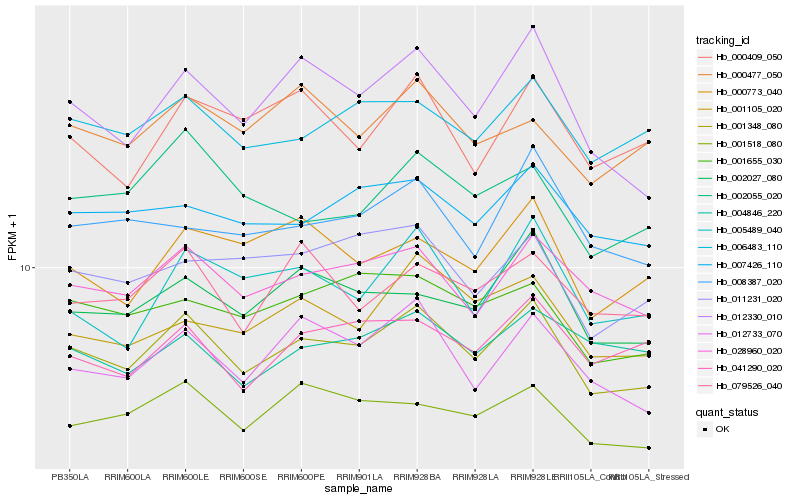
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_080 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 2 |
Hb_028960_020 |
0.0567629698 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012330_010 |
0.0612369228 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_012733_070 |
0.0655822122 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 5 |
Hb_006483_110 |
0.0680905168 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 6 |
Hb_002027_080 |
0.0691679149 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 7 |
Hb_001518_080 |
0.0703410251 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 8 |
Hb_007426_110 |
0.0712196632 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_001655_030 |
0.073454001 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001105_020 |
0.0750351017 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_005489_040 |
0.0757218648 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 12 |
Hb_011231_020 |
0.0764444834 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_041290_020 |
0.0767732959 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_079526_040 |
0.0768138402 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_000773_040 |
0.0778526594 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 16 |
Hb_008387_020 |
0.0784991708 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 17 |
Hb_000409_050 |
0.0789982464 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 18 |
Hb_002055_020 |
0.0793580947 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 19 |
Hb_000477_050 |
0.0806978316 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 20 |
Hb_004846_220 |
0.0813138666 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |