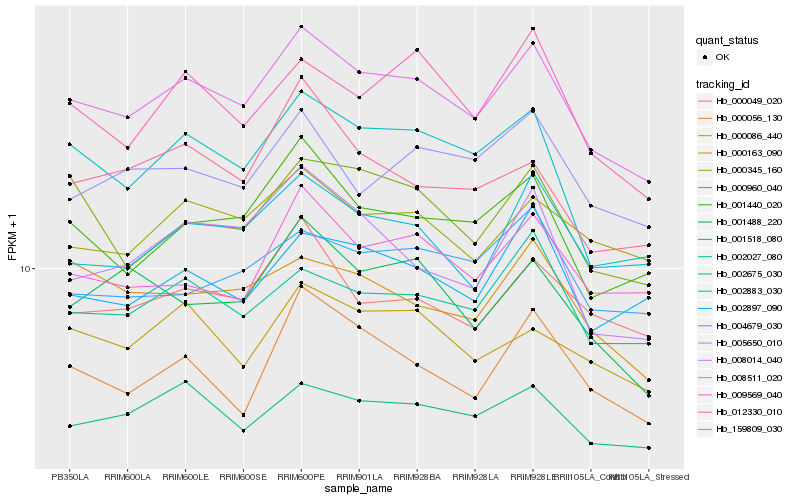
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008511_020 |
0.0 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 2 |
Hb_002675_030 |
0.0576300586 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 3 |
Hb_001440_020 |
0.0639542997 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 4 |
Hb_002027_080 |
0.0666061494 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 5 |
Hb_001518_080 |
0.0666486925 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 6 |
Hb_000049_020 |
0.0689610401 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000345_160 |
0.0690226987 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 8 |
Hb_002883_030 |
0.0693067382 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 9 |
Hb_012330_010 |
0.0706135414 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000163_090 |
0.0713805702 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 11 |
Hb_000960_040 |
0.0725949348 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 12 |
Hb_009569_040 |
0.0741409738 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 13 |
Hb_002897_090 |
0.0746713869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000056_130 |
0.0751891422 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_000086_440 |
0.0761092625 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 16 |
Hb_001488_220 |
0.0775516029 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_008014_040 |
0.077653048 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 18 |
Hb_005650_010 |
0.0776694531 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_004679_030 |
0.0787604082 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 20 |
Hb_159809_030 |
0.0798763521 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |