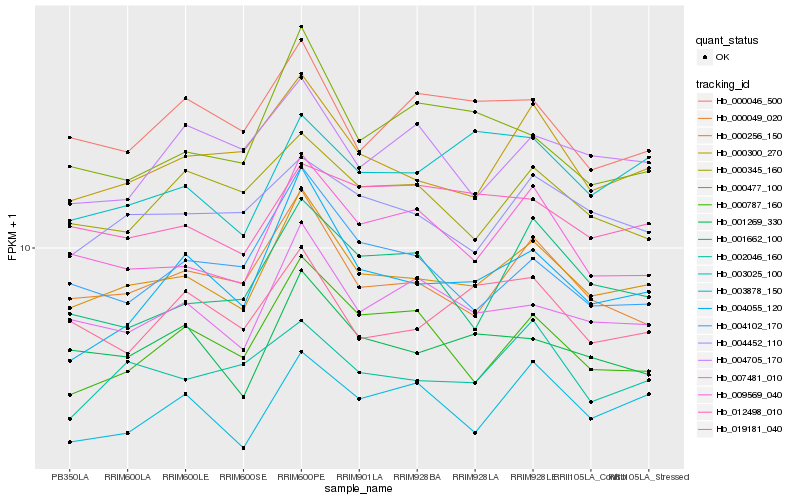
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_150 |
0.0 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 2 |
Hb_000049_020 |
0.0554533317 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_000300_270 |
0.0558917111 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_007481_010 |
0.0576421853 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 5 |
Hb_001269_330 |
0.0618743678 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 6 |
Hb_009569_040 |
0.0628845607 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 7 |
Hb_004452_110 |
0.0668817809 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003878_150 |
0.0697794457 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 9 |
Hb_004055_120 |
0.0706863932 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 10 |
Hb_002046_160 |
0.0719244329 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000046_500 |
0.073741801 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 12 |
Hb_000787_160 |
0.0746055174 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000345_160 |
0.0751326312 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 14 |
Hb_004102_170 |
0.0757392709 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 15 |
Hb_019181_040 |
0.0758742572 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 16 |
Hb_003025_100 |
0.0777155423 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001662_100 |
0.0783195715 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 18 |
Hb_012498_010 |
0.0784697801 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 19 |
Hb_000477_100 |
0.0797176819 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 20 |
Hb_004705_170 |
0.0797907256 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |