| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
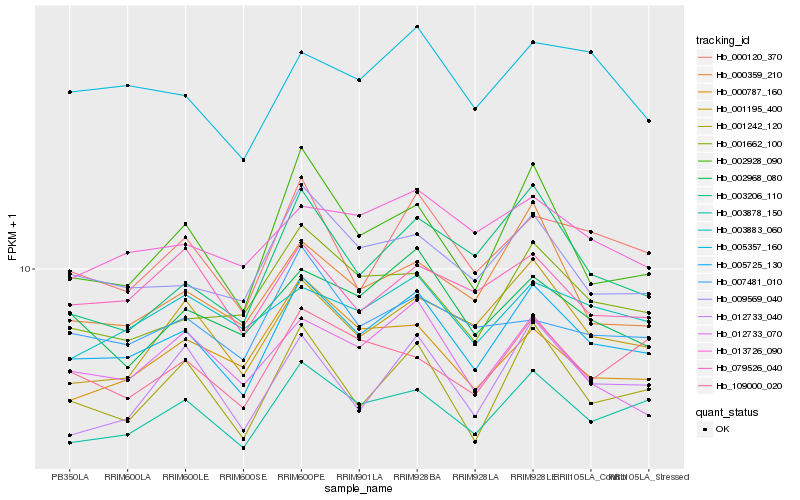
Hb_005725_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 2 |
Hb_003878_150 |
0.0534377882 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 3 |
Hb_001242_120 |
0.0636744068 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 4 |
Hb_009569_040 |
0.0646301025 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 5 |
Hb_000120_370 |
0.0671885888 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 6 |
Hb_002928_090 |
0.0692874862 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 7 |
Hb_001662_100 |
0.0720448645 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 8 |
Hb_012733_070 |
0.0763899091 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002968_080 |
0.0764398709 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 10 |
Hb_005357_160 |
0.0770007606 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 11 |
Hb_001195_400 |
0.0771010596 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 12 |
Hb_000359_210 |
0.0773347753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012733_040 |
0.0792045966 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 14 |
Hb_003883_060 |
0.0807552468 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_013726_090 |
0.0808195563 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 16 |
Hb_079526_040 |
0.0811726118 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 17 |
Hb_109000_020 |
0.0829733222 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 18 |
Hb_003206_110 |
0.0830557445 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 19 |
Hb_007481_010 |
0.0841009121 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 20 |
Hb_000787_160 |
0.0843055665 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |