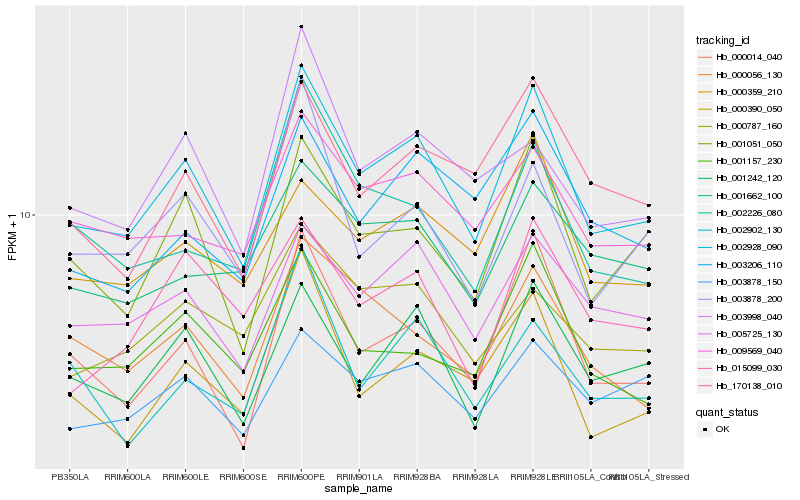
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002928_090 |
0.0 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 2 |
Hb_000014_040 |
0.066888644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001242_120 |
0.0676320386 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 4 |
Hb_001051_050 |
0.0692198148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005725_130 |
0.0692874862 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_000390_050 |
0.079232617 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 7 |
Hb_009569_040 |
0.0802116491 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 8 |
Hb_001157_230 |
0.0809146131 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 9 |
Hb_000359_210 |
0.0815844415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_170138_010 |
0.081696147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000056_130 |
0.0835257498 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 12 |
Hb_003878_200 |
0.0850163517 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 13 |
Hb_003878_150 |
0.0851938353 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_002226_080 |
0.0867192462 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003206_110 |
0.0871094882 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 16 |
Hb_001662_100 |
0.087733195 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 17 |
Hb_003998_040 |
0.0884742136 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 18 |
Hb_000787_160 |
0.089789831 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_015099_030 |
0.0904677522 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 20 |
Hb_002902_130 |
0.0912335953 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |