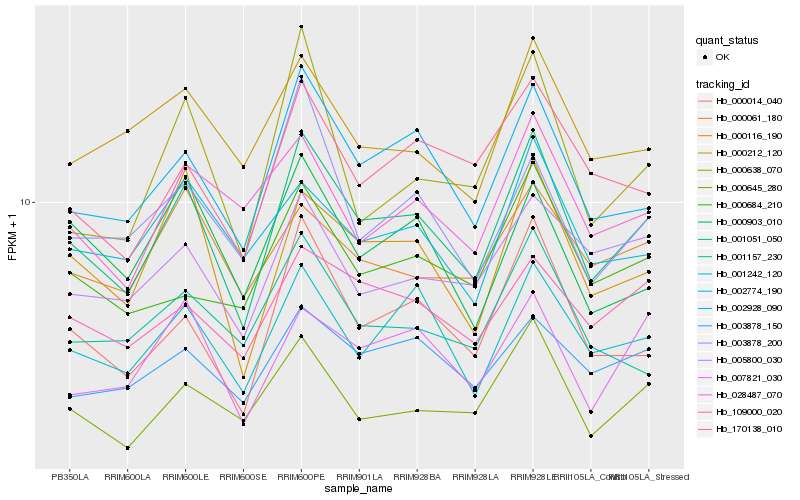
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001051_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002928_090 |
0.0692198148 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 3 |
Hb_000014_040 |
0.0761897993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001242_120 |
0.0797923723 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 5 |
Hb_000116_190 |
0.0801806839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000645_280 |
0.0844472797 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 7 |
Hb_007821_030 |
0.0900422054 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 8 |
Hb_109000_020 |
0.0949753858 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 9 |
Hb_170138_010 |
0.0951011996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003878_150 |
0.0970330321 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 11 |
Hb_002774_190 |
0.0996553624 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 12 |
Hb_000638_070 |
0.1000932806 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 13 |
Hb_000684_210 |
0.1001761567 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 14 |
Hb_005800_030 |
0.1004320517 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 15 |
Hb_000903_010 |
0.1009801575 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 16 |
Hb_028487_070 |
0.1020608626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000212_120 |
0.1024840758 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 18 |
Hb_003878_200 |
0.1030631664 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 19 |
Hb_000061_180 |
0.1034204926 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 20 |
Hb_001157_230 |
0.1038862287 |
- |
- |
Protein AFR, putative [Ricinus communis] |