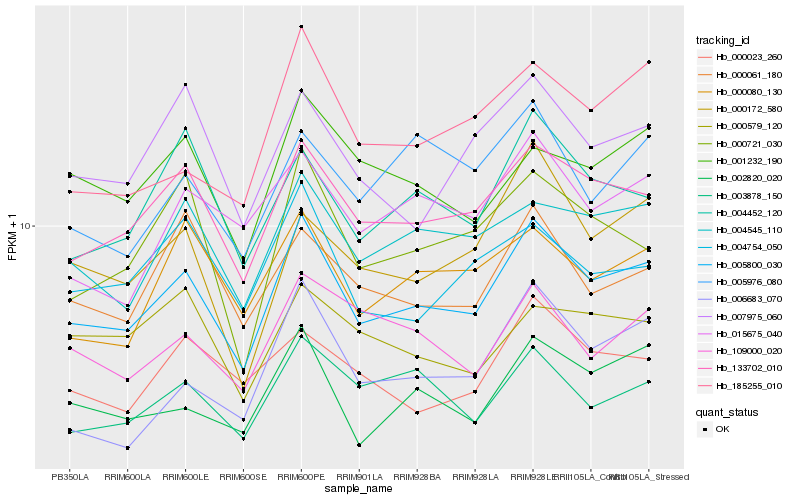
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005800_030 |
0.0 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 2 |
Hb_133702_010 |
0.0512228688 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_004545_110 |
0.0669871313 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_002820_020 |
0.0693858971 |
- |
- |
hypothetical protein VITISV_005587 [Vitis vinifera] |
| 5 |
Hb_003878_150 |
0.075227099 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_004452_120 |
0.0763975984 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 7 |
Hb_000061_180 |
0.0782480321 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 8 |
Hb_000080_130 |
0.0787422696 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 9 |
Hb_007975_060 |
0.0806442288 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001232_190 |
0.0808383899 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000721_030 |
0.0819398276 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 12 |
Hb_004754_050 |
0.082182381 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 13 |
Hb_109000_020 |
0.0824104719 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 14 |
Hb_015675_040 |
0.083468669 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 15 |
Hb_185255_010 |
0.0834787917 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 16 |
Hb_000172_580 |
0.0839526947 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000023_260 |
0.0840211276 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006683_070 |
0.0854083349 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 19 |
Hb_005976_080 |
0.08832112 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 20 |
Hb_000579_120 |
0.0896293219 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |