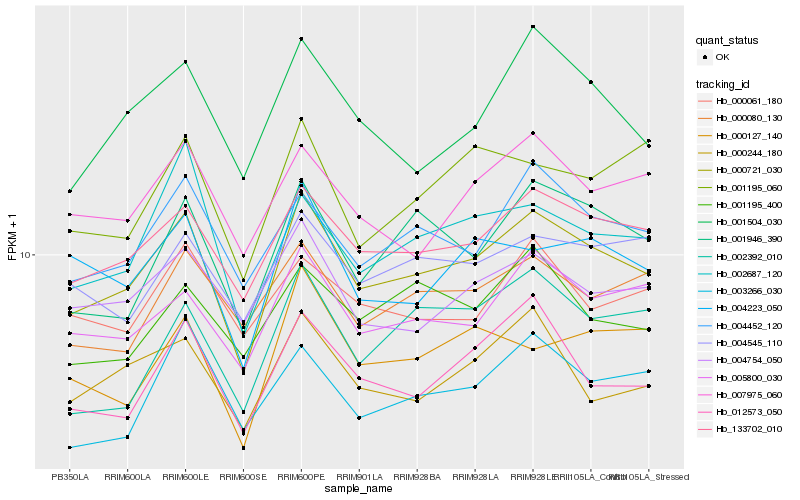
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000721_030 |
0.0 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 2 |
Hb_133702_010 |
0.0653297977 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_004452_120 |
0.0819208201 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 4 |
Hb_005800_030 |
0.0819398276 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 5 |
Hb_001946_390 |
0.0854886993 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 6 |
Hb_012573_050 |
0.0860930264 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 7 |
Hb_001504_030 |
0.0888269356 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 8 |
Hb_002687_120 |
0.0892315492 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000244_180 |
0.0898838918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004754_050 |
0.0912535318 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 11 |
Hb_004223_050 |
0.0919883633 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_007975_060 |
0.0923945663 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004545_110 |
0.0957519749 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000080_130 |
0.0963067528 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 15 |
Hb_003266_030 |
0.0975386843 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 16 |
Hb_001195_400 |
0.0997308811 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 17 |
Hb_001195_060 |
0.1001606802 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 18 |
Hb_000127_140 |
0.1004091159 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 19 |
Hb_002392_010 |
0.1013401233 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 20 |
Hb_000061_180 |
0.1015316219 |
- |
- |
exonuclease, putative [Ricinus communis] |