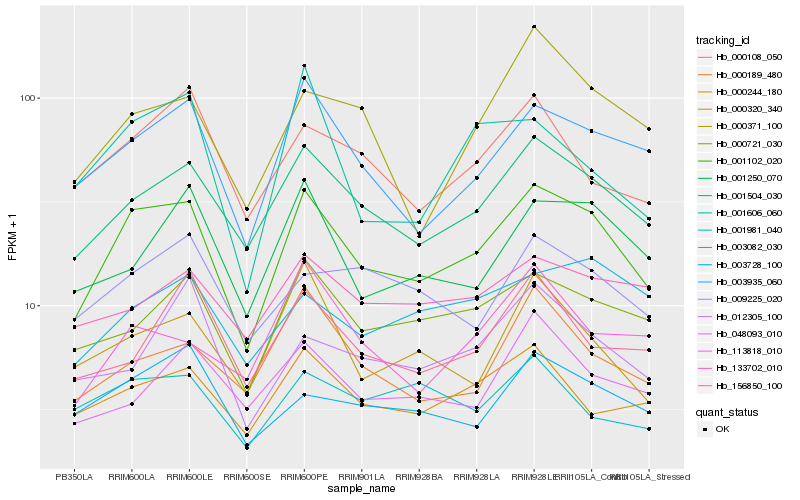
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_020 |
0.0 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_001504_030 |
0.0903898782 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 3 |
Hb_003935_060 |
0.1091061761 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 4 |
Hb_000721_030 |
0.1181633728 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 5 |
Hb_001250_070 |
0.1292516786 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 6 |
Hb_000244_180 |
0.1307613614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003728_100 |
0.1331825235 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000108_050 |
0.1338397548 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 9 |
Hb_000189_480 |
0.1365129367 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 10 |
Hb_000320_340 |
0.1374700528 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 11 |
Hb_009225_020 |
0.1392413655 |
- |
- |
PREDICTED: uncharacterized protein LOC105638232 [Jatropha curcas] |
| 12 |
Hb_000371_100 |
0.1411254644 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 13 |
Hb_048093_010 |
0.1427745078 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_133702_010 |
0.1437363322 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_156850_100 |
0.1441216891 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 16 |
Hb_012305_100 |
0.1447459334 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_003082_030 |
0.1454415471 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_113818_010 |
0.1464612452 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 19 |
Hb_001606_060 |
0.1483825125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001981_040 |
0.1489176374 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |