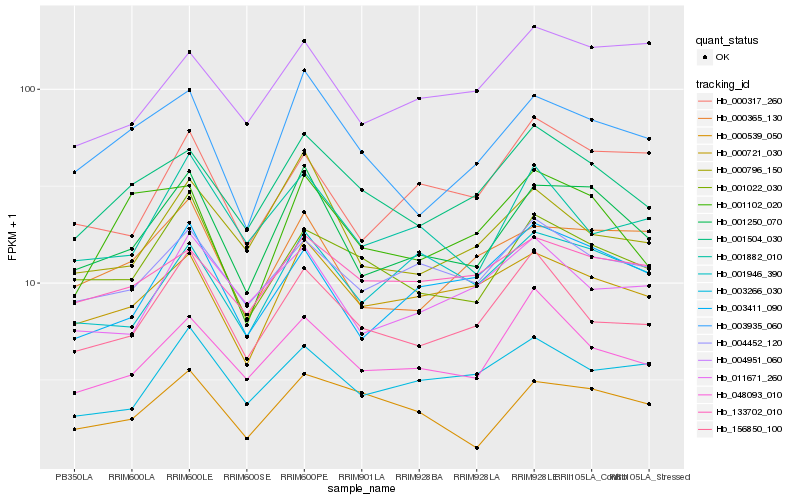
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001250_070 |
0.0 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 2 |
Hb_003935_060 |
0.1014615538 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 3 |
Hb_000721_030 |
0.1033303491 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 4 |
Hb_000365_130 |
0.1036479388 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 5 |
Hb_003411_090 |
0.1057099866 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001504_030 |
0.1096793501 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 7 |
Hb_001022_030 |
0.1108891205 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 8 |
Hb_000796_150 |
0.112058268 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 9 |
Hb_004452_120 |
0.1168653858 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 10 |
Hb_048093_010 |
0.1171571126 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_001946_390 |
0.1198079215 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 12 |
Hb_156850_100 |
0.1198251488 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 13 |
Hb_000317_260 |
0.1223889608 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_003266_030 |
0.1227356154 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 15 |
Hb_000539_050 |
0.1236869054 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 16 |
Hb_001882_010 |
0.1243007375 |
- |
- |
- |
| 17 |
Hb_011671_260 |
0.1252250471 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_133702_010 |
0.1257591398 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_004951_060 |
0.126745764 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 20 |
Hb_001102_020 |
0.1292516786 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |