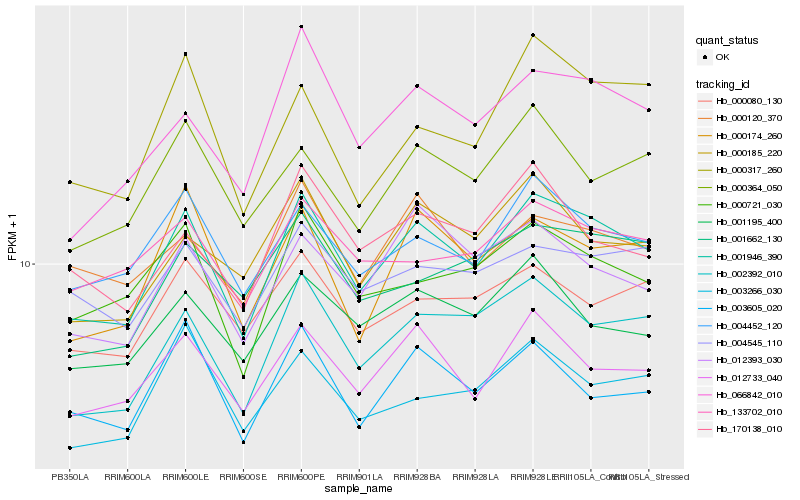
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_390 |
0.0 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 2 |
Hb_002392_010 |
0.0697205183 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 3 |
Hb_012733_040 |
0.0746944239 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 4 |
Hb_004452_120 |
0.0781538196 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 5 |
Hb_066842_010 |
0.0820511329 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000174_260 |
0.0847922391 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 7 |
Hb_000721_030 |
0.0854886993 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 8 |
Hb_000185_220 |
0.0864717936 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 9 |
Hb_001195_400 |
0.0871809565 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 10 |
Hb_012393_030 |
0.0873004268 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 11 |
Hb_000364_050 |
0.0885352809 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000080_130 |
0.0921016993 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 13 |
Hb_003605_020 |
0.0928052429 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 14 |
Hb_000317_260 |
0.0930977993 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_004545_110 |
0.0932987382 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_170138_010 |
0.0933869796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003266_030 |
0.0934779372 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 18 |
Hb_133702_010 |
0.0938218324 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000120_370 |
0.0940831937 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 20 |
Hb_001662_130 |
0.0952731503 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |