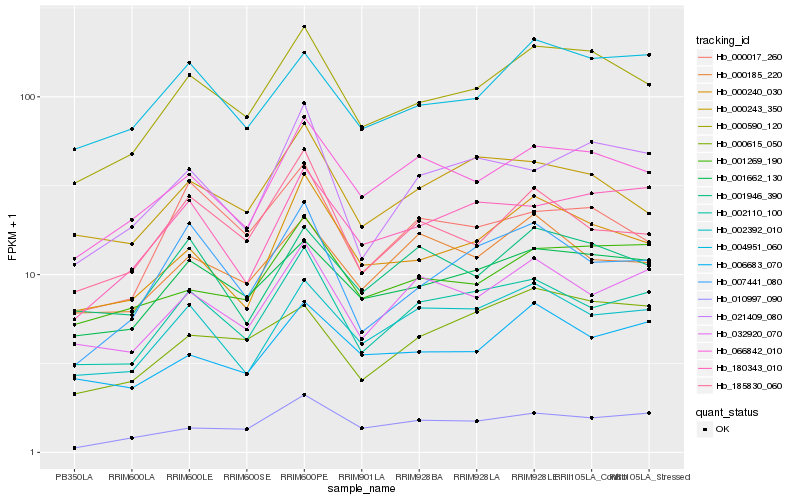
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000590_120 |
0.0 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 2 |
Hb_001662_130 |
0.0746561931 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000240_030 |
0.0803966979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_066842_010 |
0.0841988804 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001269_190 |
0.096223647 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 6 |
Hb_002110_100 |
0.0965399253 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004951_060 |
0.0983884221 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 8 |
Hb_002392_010 |
0.1040111144 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 9 |
Hb_180343_010 |
0.1043367147 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 10 |
Hb_010997_090 |
0.1062612693 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 11 |
Hb_000243_350 |
0.1069163616 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 12 |
Hb_006683_070 |
0.107809626 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 13 |
Hb_000017_260 |
0.1080439646 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 14 |
Hb_007441_080 |
0.1084852173 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 15 |
Hb_001946_390 |
0.1085827694 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 16 |
Hb_000615_050 |
0.1088651888 |
- |
- |
Potassium transporter 11 family protein [Populus trichocarpa] |
| 17 |
Hb_185830_060 |
0.1098467896 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 18 |
Hb_032920_070 |
0.1112048879 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 19 |
Hb_021409_080 |
0.11248383 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000185_220 |
0.1146811774 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |