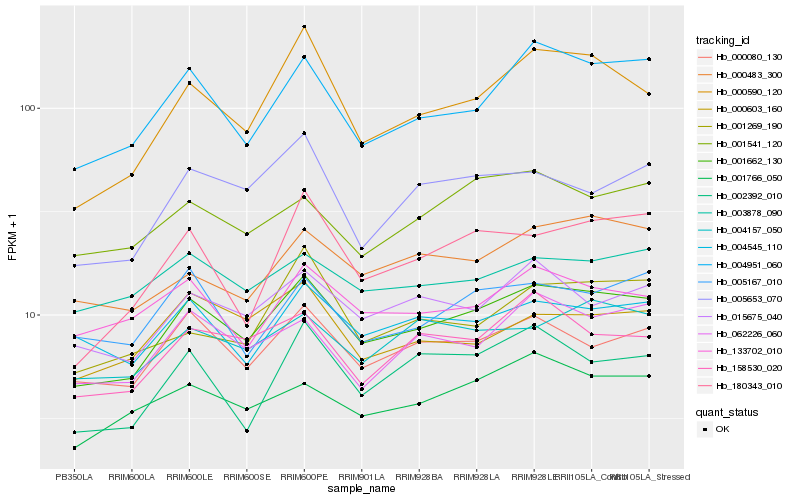
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_130 |
0.0 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004951_060 |
0.0709683782 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 3 |
Hb_000080_130 |
0.0724684413 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 4 |
Hb_000590_120 |
0.0746561931 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 5 |
Hb_015675_040 |
0.0786583381 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 6 |
Hb_001766_050 |
0.0823702194 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 7 |
Hb_005653_070 |
0.0824984684 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 8 |
Hb_158530_020 |
0.0834095575 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_062226_060 |
0.0851847579 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 10 |
Hb_004157_050 |
0.0869860672 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000483_300 |
0.0873315002 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 12 |
Hb_002392_010 |
0.0878975029 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 13 |
Hb_004545_110 |
0.0890929089 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_180343_010 |
0.0891278604 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 15 |
Hb_133702_010 |
0.0895152052 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000603_160 |
0.0896874859 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 17 |
Hb_005167_010 |
0.0899993917 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003878_090 |
0.0909201881 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 19 |
Hb_001541_120 |
0.0910438589 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 20 |
Hb_001269_190 |
0.0931483053 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |