| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
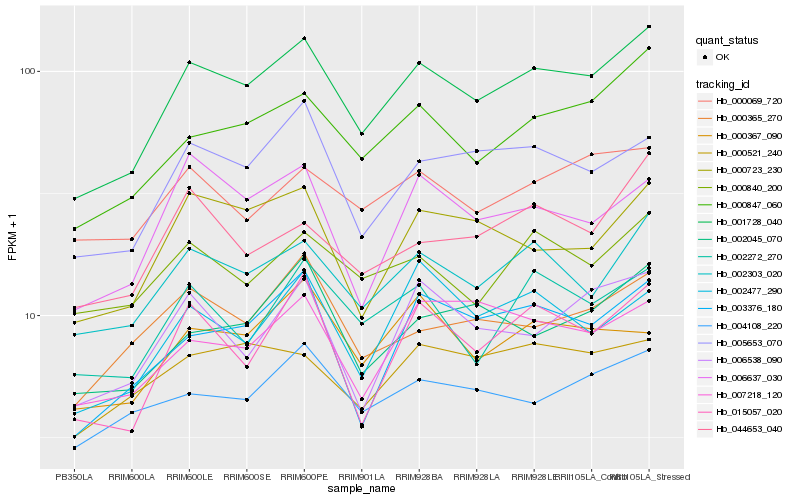
Hb_001728_040 |
0.0 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 2 |
Hb_002303_020 |
0.0679018082 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 3 |
Hb_000847_060 |
0.0694365395 |
- |
- |
hypothetical protein [Jatropha curcas] |
| 4 |
Hb_005653_070 |
0.0758790521 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 5 |
Hb_000723_230 |
0.0838616518 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 6 |
Hb_015057_020 |
0.0845783015 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 7 |
Hb_003376_180 |
0.0870849293 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 8 |
Hb_002272_270 |
0.0891800869 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 9 |
Hb_006637_030 |
0.089432167 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 10 |
Hb_000365_270 |
0.0898234395 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 11 |
Hb_002045_070 |
0.0901586695 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 12 |
Hb_000840_200 |
0.0905972311 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004108_220 |
0.0907818768 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002477_290 |
0.0916176408 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_006538_090 |
0.093738886 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 16 |
Hb_000367_090 |
0.0937422394 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 17 |
Hb_000521_240 |
0.0944129879 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007218_120 |
0.0944338982 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000069_720 |
0.0945774328 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 20 |
Hb_044653_040 |
0.0948903006 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |