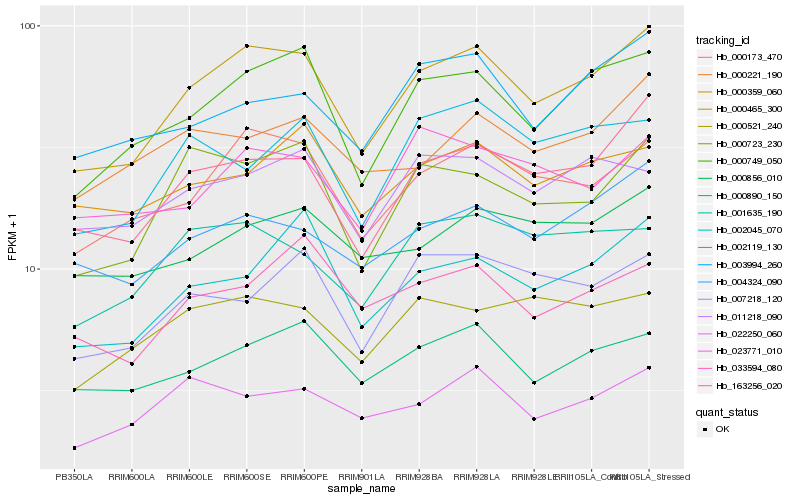
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_300 |
0.0 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 2 |
Hb_000749_050 |
0.0645803821 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 3 |
Hb_000173_470 |
0.0655991153 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_163256_020 |
0.0669268354 |
- |
- |
fructokinase [Manihot esculenta] |
| 5 |
Hb_000723_230 |
0.0838949424 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 6 |
Hb_000890_150 |
0.0862315143 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 7 |
Hb_002045_070 |
0.0881251543 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 8 |
Hb_004324_090 |
0.088145221 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 9 |
Hb_022250_060 |
0.0937861908 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001635_190 |
0.094030529 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003994_260 |
0.0948922251 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 12 |
Hb_007218_120 |
0.0950678342 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 13 |
Hb_033594_080 |
0.0974723481 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 14 |
Hb_000521_240 |
0.0984048989 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011218_090 |
0.0986167098 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 16 |
Hb_000856_010 |
0.0987821315 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 17 |
Hb_023771_010 |
0.0999292101 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 18 |
Hb_002119_130 |
0.1009948024 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 19 |
Hb_000359_060 |
0.1011239423 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 20 |
Hb_000221_190 |
0.1015350041 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |