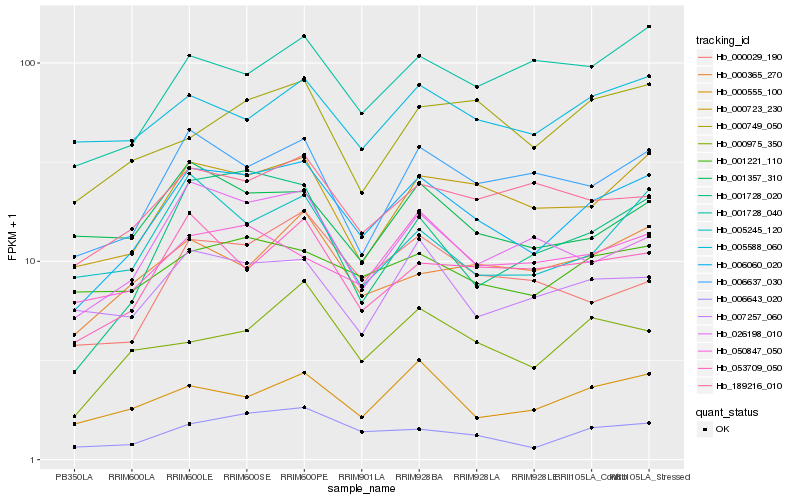
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006060_020 |
0.0 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000723_230 |
0.0918458546 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 3 |
Hb_006643_020 |
0.093074448 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 4 |
Hb_006637_030 |
0.1047579727 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 5 |
Hb_026198_010 |
0.1094046743 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 6 |
Hb_053709_050 |
0.1105950188 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000555_100 |
0.1134061947 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 8 |
Hb_000975_350 |
0.1153973625 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 9 |
Hb_000365_270 |
0.1167449224 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 10 |
Hb_001728_020 |
0.1170075591 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 11 |
Hb_001728_040 |
0.1171037273 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 12 |
Hb_005245_120 |
0.1173886373 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 13 |
Hb_189216_010 |
0.118090943 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_110 |
0.1183806665 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 15 |
Hb_000029_190 |
0.1208779987 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 16 |
Hb_005588_060 |
0.1212246288 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 17 |
Hb_007257_060 |
0.1213690765 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 18 |
Hb_050847_050 |
0.1217513129 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001357_310 |
0.1235229679 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000749_050 |
0.1262452396 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |