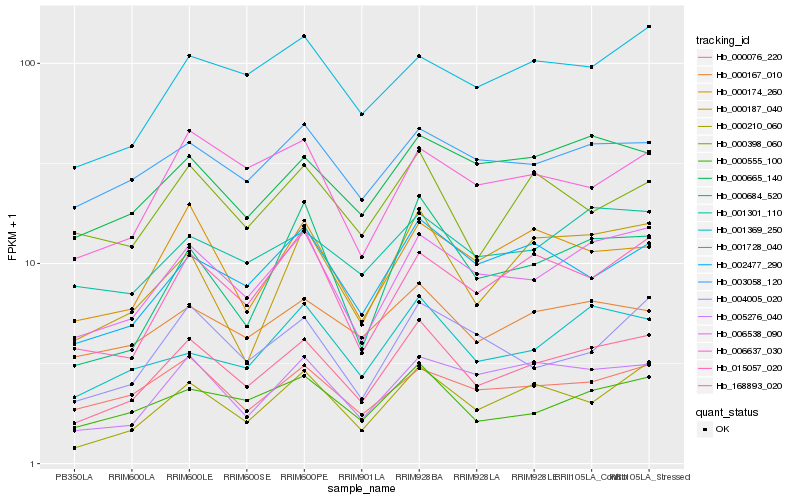
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168893_020 |
0.0 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 2 |
Hb_006538_090 |
0.0710724579 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 3 |
Hb_000210_060 |
0.087489064 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 4 |
Hb_000555_100 |
0.0894220671 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 5 |
Hb_001369_250 |
0.0998304774 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002477_290 |
0.1000478525 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_005276_040 |
0.1023408857 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 8 |
Hb_000187_040 |
0.1027854689 |
- |
- |
PREDICTED: pyridoxal kinase isoform X5 [Vitis vinifera] |
| 9 |
Hb_015057_020 |
0.1037522121 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 10 |
Hb_000684_520 |
0.1040880255 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 11 |
Hb_001728_040 |
0.1063418497 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 12 |
Hb_000167_010 |
0.1068868108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004005_020 |
0.1070851969 |
- |
- |
PREDICTED: nudix hydrolase 20, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000665_140 |
0.1071952448 |
- |
- |
PREDICTED: protein MEMO1 [Jatropha curcas] |
| 15 |
Hb_001301_110 |
0.109474003 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 16 |
Hb_000174_260 |
0.1097361453 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 17 |
Hb_006637_030 |
0.1109506924 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 18 |
Hb_000076_220 |
0.111454915 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_003058_120 |
0.1149150275 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 20 |
Hb_000398_060 |
0.1174271456 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |