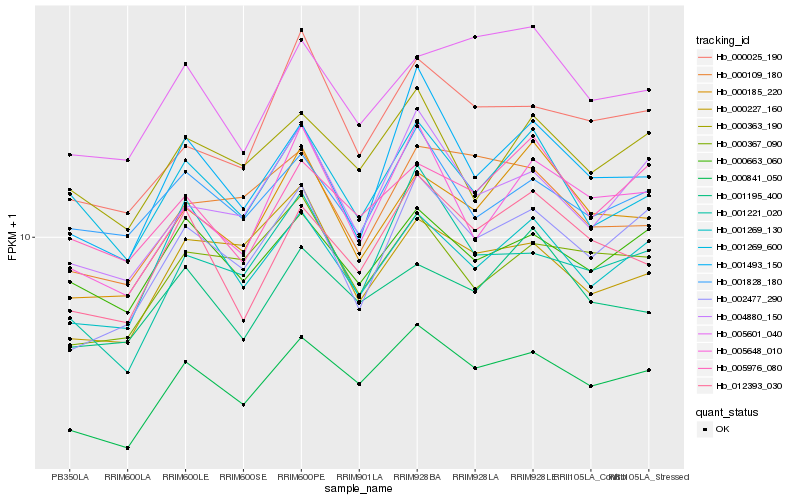
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000841_050 |
0.0 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 2 |
Hb_012393_030 |
0.0725320064 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 3 |
Hb_002477_290 |
0.0762846633 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001221_020 |
0.0768346827 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 5 |
Hb_004880_150 |
0.0791969013 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 6 |
Hb_000663_060 |
0.0811221296 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 7 |
Hb_001269_130 |
0.0816949876 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 8 |
Hb_000025_190 |
0.0839590338 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_000367_090 |
0.0846187028 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 10 |
Hb_005648_010 |
0.0857157059 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 11 |
Hb_000185_220 |
0.0860570623 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 12 |
Hb_000363_190 |
0.08606641 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001493_150 |
0.0892145838 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 14 |
Hb_005976_080 |
0.09151311 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 15 |
Hb_005601_040 |
0.0916793656 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_001828_180 |
0.0918614212 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 17 |
Hb_000109_180 |
0.0922319876 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 18 |
Hb_001269_600 |
0.0926287155 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_001195_400 |
0.0926930335 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 20 |
Hb_000227_160 |
0.0943869093 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |