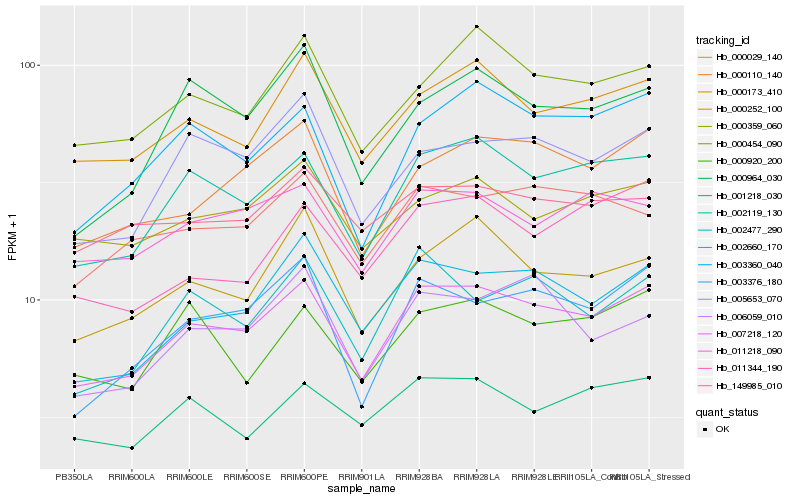
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007218_120 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002119_130 |
0.0491706318 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 3 |
Hb_000173_410 |
0.0674503439 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 4 |
Hb_002660_170 |
0.0676461242 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_000110_140 |
0.0725906702 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 6 |
Hb_003360_040 |
0.0733581485 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 7 |
Hb_003376_180 |
0.0747669543 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 8 |
Hb_000454_090 |
0.0748575307 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 9 |
Hb_000252_100 |
0.0772806068 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 10 |
Hb_005653_070 |
0.07788168 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 11 |
Hb_002477_290 |
0.0787358318 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_011218_090 |
0.0797476911 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 13 |
Hb_000029_140 |
0.0811075523 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 14 |
Hb_001218_030 |
0.0817229031 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 15 |
Hb_006059_010 |
0.0828840283 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 16 |
Hb_149985_010 |
0.0833063142 |
- |
- |
PREDICTED: transcription initiation factor IIB-2 [Cucumis sativus] |
| 17 |
Hb_000359_060 |
0.0849624497 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 18 |
Hb_000920_200 |
0.0854570542 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_011344_190 |
0.0855256472 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 20 |
Hb_000964_030 |
0.0871195722 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |