| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
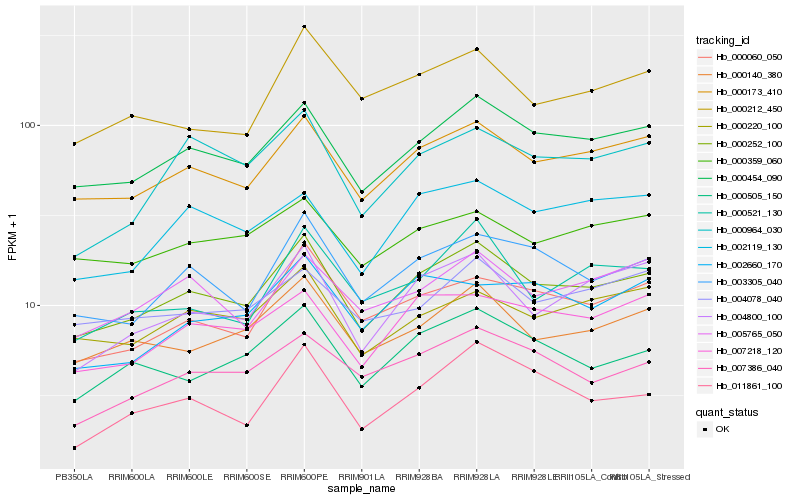
Hb_000252_100 |
0.0 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 2 |
Hb_000454_090 |
0.0427061091 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 3 |
Hb_000173_410 |
0.0455831668 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 4 |
Hb_003305_040 |
0.067262291 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 5 |
Hb_000140_380 |
0.0694803675 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 6 |
Hb_000060_050 |
0.0760272907 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 7 |
Hb_000505_150 |
0.0766337634 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 8 |
Hb_007218_120 |
0.0772806068 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 9 |
Hb_004800_100 |
0.0775322698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000220_100 |
0.0799860697 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 11 |
Hb_007386_040 |
0.0816538447 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000964_030 |
0.0839907002 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 13 |
Hb_005765_050 |
0.0843268632 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4-like [Populus euphratica] |
| 14 |
Hb_002119_130 |
0.0859695678 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 15 |
Hb_000359_060 |
0.0861232096 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 16 |
Hb_000212_450 |
0.0878196521 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_000521_130 |
0.0895781923 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_004078_040 |
0.0905843066 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 19 |
Hb_002660_170 |
0.0944771858 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_011861_100 |
0.0953867953 |
- |
- |
amino acid transporter, putative [Ricinus communis] |