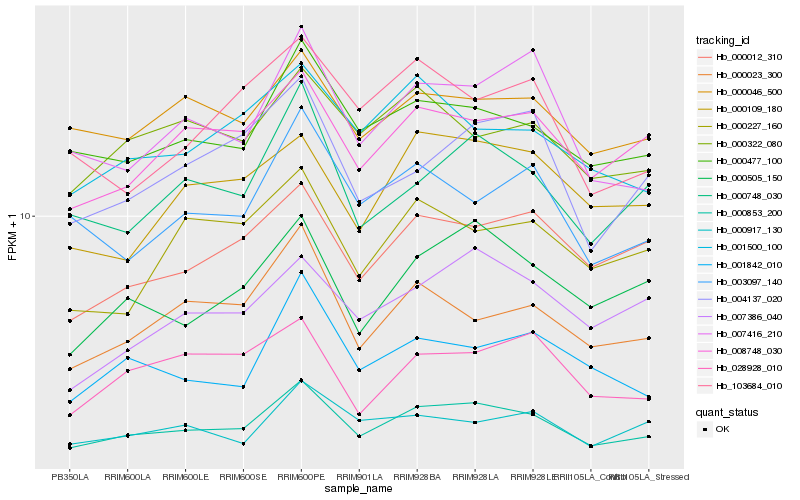
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 2 |
Hb_000477_100 |
0.082967383 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 3 |
Hb_000505_150 |
0.0836513733 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 4 |
Hb_008748_030 |
0.0842048294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000748_030 |
0.0850222351 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000012_310 |
0.0881780002 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_007416_210 |
0.0892333755 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 8 |
Hb_000023_300 |
0.0928159666 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 9 |
Hb_004137_020 |
0.0951588526 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000109_180 |
0.0960889476 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 11 |
Hb_007386_040 |
0.0988334241 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_001500_100 |
0.0993029047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003097_140 |
0.0997647155 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 14 |
Hb_000322_080 |
0.0998443401 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 15 |
Hb_001842_010 |
0.1002704219 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000046_500 |
0.101848566 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 17 |
Hb_000227_160 |
0.1028741659 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 18 |
Hb_000917_130 |
0.1046607064 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 19 |
Hb_028928_010 |
0.1051930367 |
- |
- |
hypothetical protein CISIN_1g0030132mg, partial [Citrus sinensis] |
| 20 |
Hb_103684_010 |
0.1053298943 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |