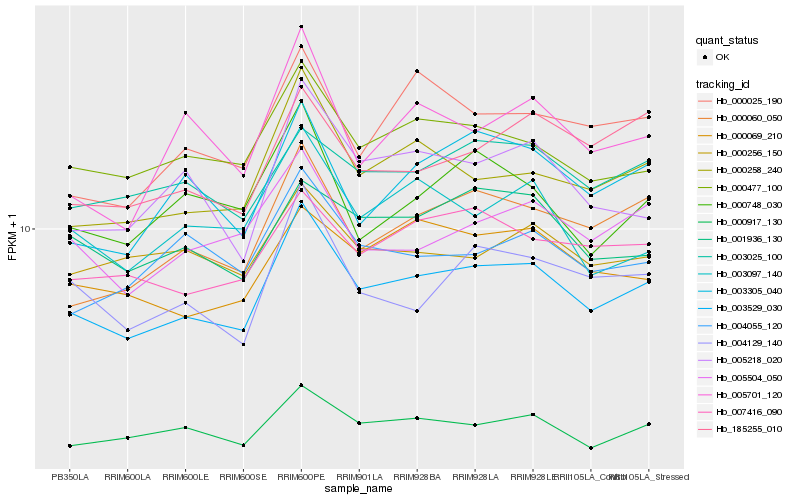
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003529_030 |
0.0 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 2 |
Hb_000258_240 |
0.0722838705 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 3 |
Hb_000917_130 |
0.0763407126 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 4 |
Hb_000477_100 |
0.0773591663 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 5 |
Hb_000060_050 |
0.0798271773 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 6 |
Hb_004129_140 |
0.0838131573 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 7 |
Hb_005504_050 |
0.0865210469 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 8 |
Hb_007416_090 |
0.0879706743 |
- |
- |
UDP-sugar transporter, putative [Ricinus communis] |
| 9 |
Hb_005218_020 |
0.0905377175 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 10 |
Hb_003305_040 |
0.0906745214 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 11 |
Hb_003097_140 |
0.0935146017 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 12 |
Hb_000069_210 |
0.0936945492 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 13 |
Hb_000025_190 |
0.0942774933 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 14 |
Hb_000748_030 |
0.0947878605 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_001936_130 |
0.0960547114 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 16 |
Hb_004055_120 |
0.0972567855 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 17 |
Hb_005701_120 |
0.0974116488 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 18 |
Hb_003025_100 |
0.097422357 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000256_150 |
0.0977593919 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 20 |
Hb_185255_010 |
0.0980924886 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |