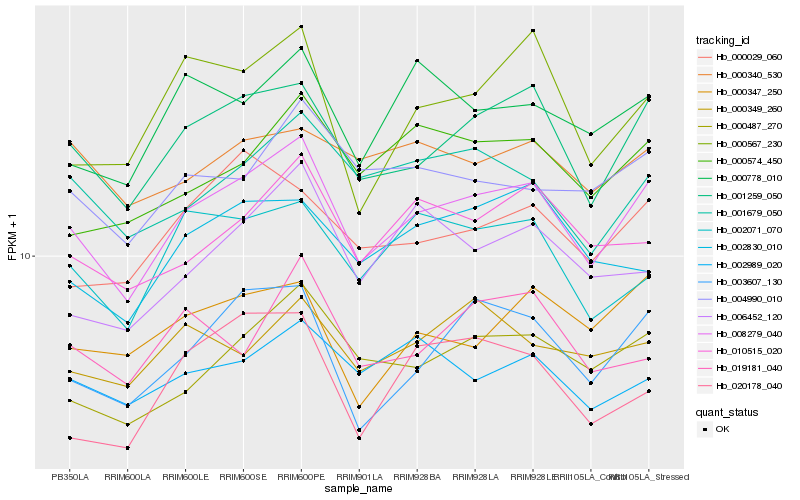
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008279_040 |
0.0 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_001259_050 |
0.0411421109 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_003607_130 |
0.0594013757 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000487_270 |
0.0637715089 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 5 |
Hb_010515_020 |
0.0683970724 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 6 |
Hb_001679_050 |
0.0691988561 |
- |
- |
PREDICTED: exocyst complex component EXO84B [Jatropha curcas] |
| 7 |
Hb_002071_070 |
0.0721584376 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000778_010 |
0.0732895253 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 9 |
Hb_004990_010 |
0.0739785881 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000029_060 |
0.0747302149 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 11 |
Hb_000574_450 |
0.0748659829 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 12 |
Hb_000567_230 |
0.0760934785 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 13 |
Hb_002989_020 |
0.0764358216 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002830_010 |
0.0768616542 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000347_250 |
0.0769177544 |
- |
- |
PREDICTED: uncharacterized protein LOC105133971 [Populus euphratica] |
| 16 |
Hb_006452_120 |
0.0784634936 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 17 |
Hb_020178_040 |
0.079078557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_019181_040 |
0.0794190952 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 19 |
Hb_000340_530 |
0.0798539168 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 20 |
Hb_000349_260 |
0.0798728858 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |