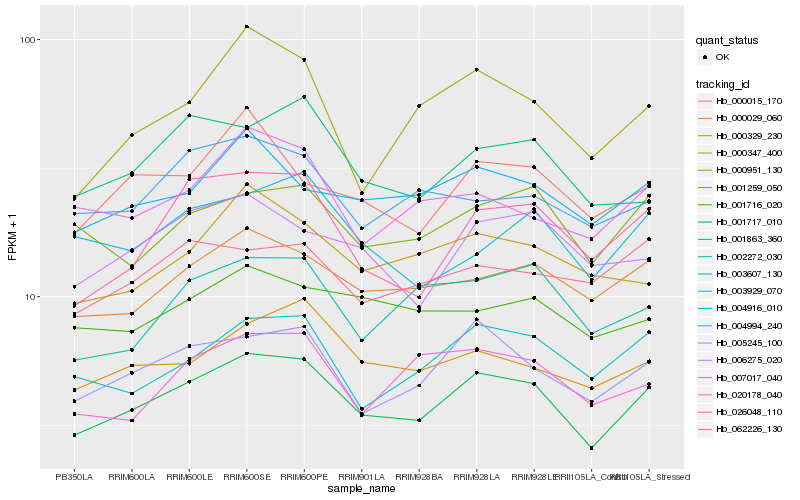
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001717_010 |
0.0 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 2 |
Hb_005245_100 |
0.0666496705 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000029_060 |
0.0748218819 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 4 |
Hb_026048_110 |
0.0801952917 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 5 |
Hb_000329_230 |
0.0803557594 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 6 |
Hb_003607_130 |
0.0807875175 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_004916_010 |
0.0825597084 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 8 |
Hb_001259_050 |
0.0835324709 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 9 |
Hb_002272_030 |
0.0856188446 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 10 |
Hb_001863_360 |
0.086556149 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 11 |
Hb_006275_020 |
0.0874593849 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 12 |
Hb_000015_170 |
0.0879841359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007017_040 |
0.0888900982 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 14 |
Hb_020178_040 |
0.0892527071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000347_400 |
0.0894156552 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004994_240 |
0.0898276704 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 17 |
Hb_000951_130 |
0.0910062878 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 18 |
Hb_003929_070 |
0.0924387011 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 19 |
Hb_001716_020 |
0.0926080618 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 20 |
Hb_062226_130 |
0.0927771523 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |