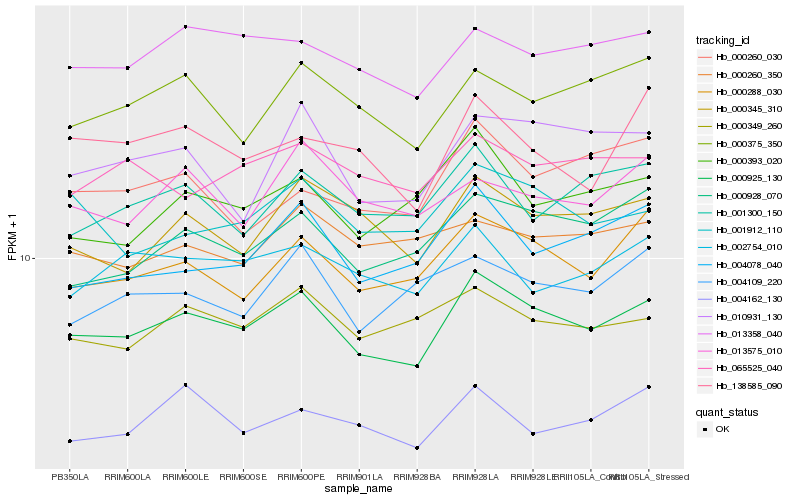
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000925_130 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000288_030 |
0.0656922124 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 3 |
Hb_013358_040 |
0.0712668997 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 4 |
Hb_000928_070 |
0.0716026006 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000349_260 |
0.0727480625 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 6 |
Hb_000345_310 |
0.073929909 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 7 |
Hb_000375_350 |
0.0759249056 |
- |
- |
PREDICTED: uncharacterized protein LOC105641632 [Jatropha curcas] |
| 8 |
Hb_001912_110 |
0.0767215574 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_002754_010 |
0.0772569855 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 10 |
Hb_010931_130 |
0.0779451768 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 11 |
Hb_004078_040 |
0.078575357 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 12 |
Hb_000393_020 |
0.0791498494 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_001300_150 |
0.0797435857 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 14 |
Hb_065525_040 |
0.0812834853 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 15 |
Hb_004109_220 |
0.0839043484 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 16 |
Hb_138585_090 |
0.0840402916 |
- |
- |
Long chain base2 isoform 2 [Theobroma cacao] |
| 17 |
Hb_004162_130 |
0.0847863906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013575_010 |
0.0848724571 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 19 |
Hb_000260_350 |
0.085923979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000260_030 |
0.0860438437 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |