| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
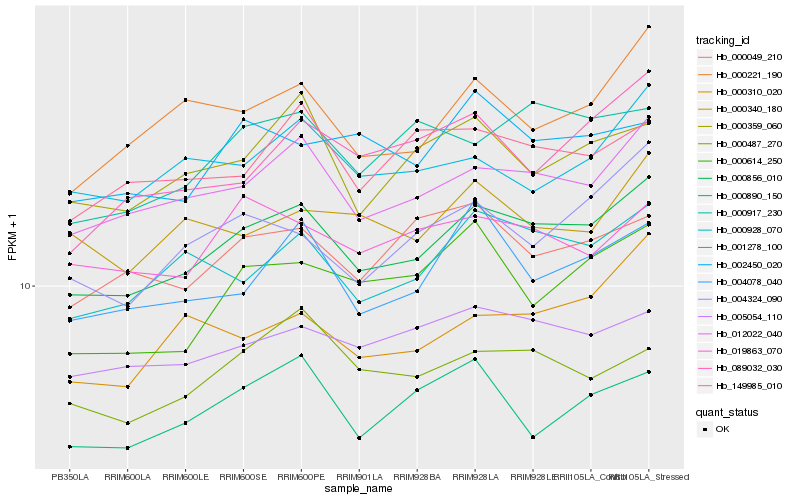
Hb_000856_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 2 |
Hb_012022_040 |
0.0437258192 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 3 |
Hb_000928_070 |
0.0579886671 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_005054_110 |
0.0656278315 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 5 |
Hb_004078_040 |
0.0682826213 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 6 |
Hb_000221_190 |
0.0686759551 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 7 |
Hb_000359_060 |
0.06991754 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 8 |
Hb_000917_230 |
0.0719211971 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 9 |
Hb_004324_090 |
0.0720207942 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 10 |
Hb_149985_010 |
0.0728838334 |
- |
- |
PREDICTED: transcription initiation factor IIB-2 [Cucumis sativus] |
| 11 |
Hb_000487_270 |
0.0732812539 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 12 |
Hb_000049_210 |
0.0738864246 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 13 |
Hb_001278_100 |
0.0747523758 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 14 |
Hb_019863_070 |
0.0752876096 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000310_020 |
0.0758035906 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 16 |
Hb_000614_250 |
0.0764210414 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 17 |
Hb_002450_020 |
0.0773458437 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 18 |
Hb_089032_030 |
0.0779324755 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000890_150 |
0.0779713469 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 20 |
Hb_000340_180 |
0.0786853265 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of NFKB 1 [Jatropha curcas] |