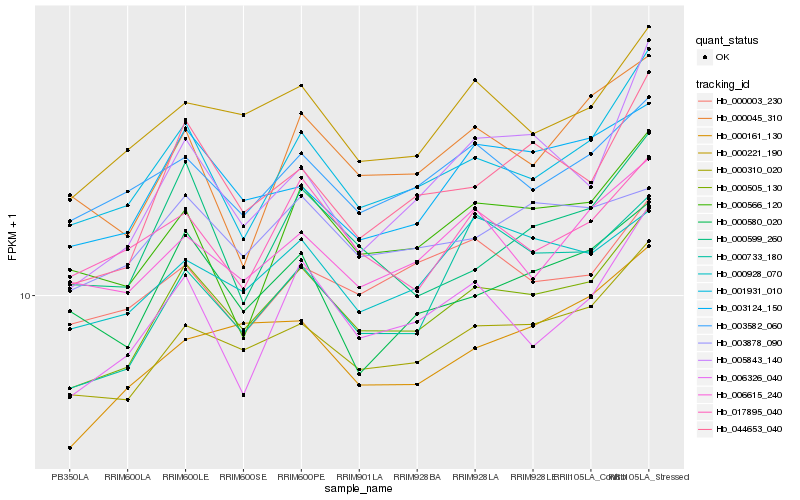
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000505_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641262 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000310_020 |
0.0450225731 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 3 |
Hb_001931_010 |
0.0571900701 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 4 |
Hb_000221_190 |
0.0605136825 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 5 |
Hb_044653_040 |
0.0609042572 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 6 |
Hb_003124_150 |
0.0699567556 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 7 |
Hb_006326_040 |
0.0711081071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003582_060 |
0.0735792061 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000566_120 |
0.076100572 |
- |
- |
PREDICTED: uncharacterized protein LOC105123035 isoform X1 [Populus euphratica] |
| 10 |
Hb_000161_130 |
0.0765434274 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006615_240 |
0.0767161772 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 12 |
Hb_000928_070 |
0.0781416883 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000580_020 |
0.0783898006 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_017895_040 |
0.0792842047 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 15 |
Hb_003878_090 |
0.0793912048 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 16 |
Hb_000599_260 |
0.0795188063 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 17 |
Hb_000733_180 |
0.0803279112 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 18 |
Hb_000003_230 |
0.0835891592 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005843_140 |
0.0843870655 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000045_310 |
0.0844554218 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 2 [Jatropha curcas] |