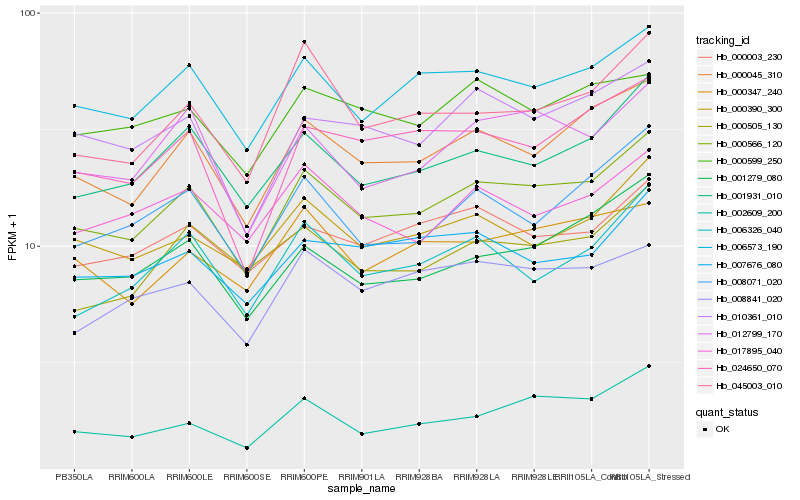
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000566_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105123035 isoform X1 [Populus euphratica] |
| 2 |
Hb_000045_310 |
0.0461497194 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 2 [Jatropha curcas] |
| 3 |
Hb_006573_190 |
0.0534554127 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001931_010 |
0.060020666 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 5 |
Hb_012799_170 |
0.062872878 |
- |
- |
PREDICTED: protein GLC8 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010361_010 |
0.0661558358 |
- |
- |
PREDICTED: ras-related protein RABH1b [Jatropha curcas] |
| 7 |
Hb_000390_300 |
0.0672848497 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 8 |
Hb_001279_080 |
0.0683310456 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105140180 isoform X1 [Populus euphratica] |
| 9 |
Hb_008071_020 |
0.0699437083 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC1 [Jatropha curcas] |
| 10 |
Hb_024650_070 |
0.0718495185 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A-like [Jatropha curcas] |
| 11 |
Hb_006326_040 |
0.071899028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002609_200 |
0.0722892259 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000003_230 |
0.0741232178 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_045003_010 |
0.0742334542 |
- |
- |
Stomatin-1, putative [Ricinus communis] |
| 15 |
Hb_008841_020 |
0.0746611886 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000347_240 |
0.0748064505 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 17 |
Hb_000505_130 |
0.076100572 |
- |
- |
PREDICTED: uncharacterized protein LOC105641262 isoform X1 [Jatropha curcas] |
| 18 |
Hb_017895_040 |
0.0764452656 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 19 |
Hb_000599_250 |
0.0771380863 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 20 |
Hb_007676_080 |
0.0773716925 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |