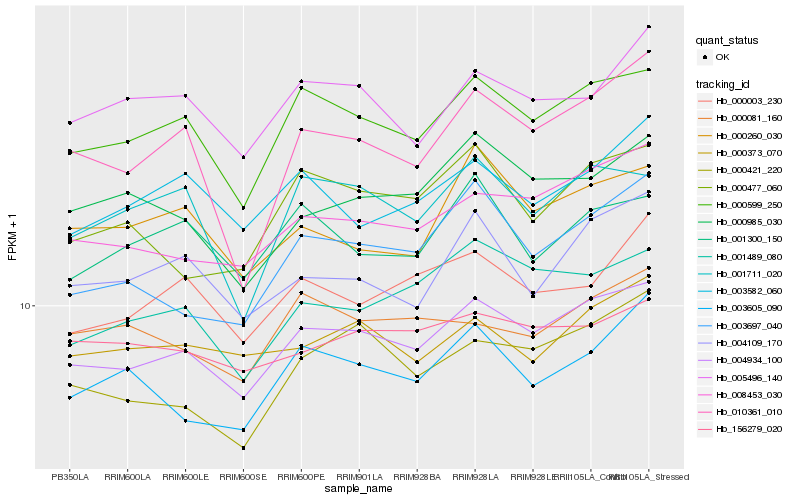
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004934_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000599_250 |
0.035978497 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 3 |
Hb_008453_030 |
0.0447512339 |
- |
- |
Thioredoxin superfamily protein [Theobroma cacao] |
| 4 |
Hb_001489_080 |
0.058984997 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000421_220 |
0.0631461989 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_003697_040 |
0.0631636512 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 7 |
Hb_000985_030 |
0.0646785133 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 8 |
Hb_000373_070 |
0.0647461971 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_004109_170 |
0.0649975309 |
- |
- |
PREDICTED: probable ribokinase [Jatropha curcas] |
| 10 |
Hb_000003_230 |
0.0654996435 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000477_060 |
0.0663225164 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 12 |
Hb_005496_140 |
0.0663751866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_156279_020 |
0.0666010634 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 14 |
Hb_001711_020 |
0.0682026062 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_000081_160 |
0.0683541402 |
- |
- |
PREDICTED: uncharacterized protein At4g17910 [Jatropha curcas] |
| 16 |
Hb_003605_090 |
0.0689903195 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632419 [Jatropha curcas] |
| 17 |
Hb_010361_010 |
0.0690941546 |
- |
- |
PREDICTED: ras-related protein RABH1b [Jatropha curcas] |
| 18 |
Hb_003582_060 |
0.0691191773 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000260_030 |
0.0701416545 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_001300_150 |
0.0707808615 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |