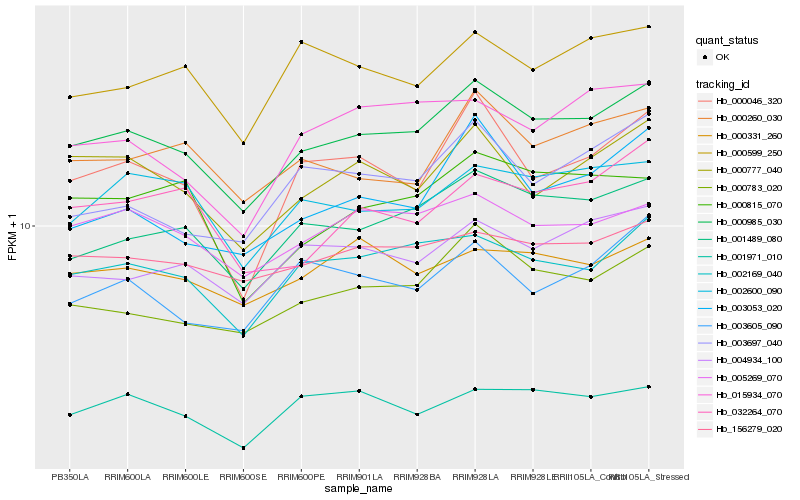
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000985_030 |
0.0 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 2 |
Hb_001489_080 |
0.0501455741 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_002169_040 |
0.0525375437 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_000783_020 |
0.0544385365 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase SDP6, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000777_040 |
0.0630061586 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 6 |
Hb_005269_070 |
0.0632220165 |
- |
- |
PREDICTED: translation factor GUF1 homolog, organellar chromatophore [Jatropha curcas] |
| 7 |
Hb_004934_100 |
0.0646785133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_156279_020 |
0.0648591345 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 9 |
Hb_000260_030 |
0.067274498 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 10 |
Hb_002600_090 |
0.0677573248 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003053_020 |
0.0704188668 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 12 |
Hb_001971_010 |
0.0724002505 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 13 |
Hb_000046_320 |
0.0735074791 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_000331_260 |
0.0742269705 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003697_040 |
0.0752424201 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 16 |
Hb_000599_250 |
0.0753907952 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 17 |
Hb_000815_070 |
0.0754126894 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 18 |
Hb_032264_070 |
0.0760798485 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003605_090 |
0.0762077052 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632419 [Jatropha curcas] |
| 20 |
Hb_015934_070 |
0.0762220828 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |