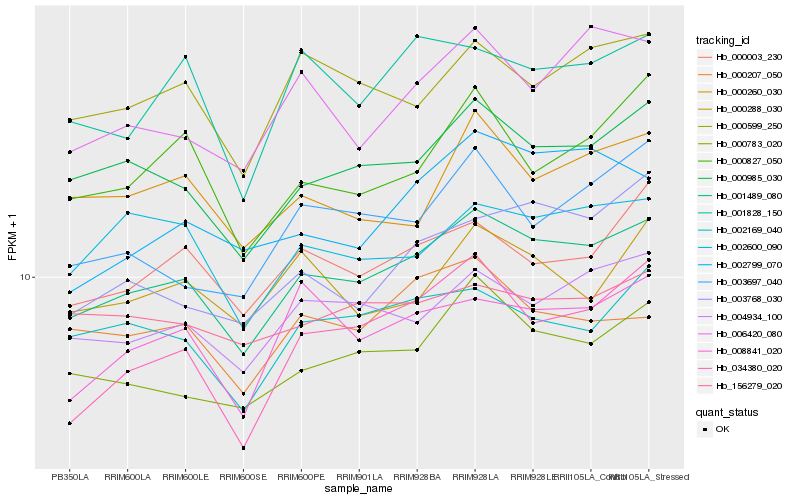
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001489_080 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000985_030 |
0.0501455741 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 3 |
Hb_004934_100 |
0.058984997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000783_020 |
0.0604448318 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase SDP6, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000599_250 |
0.0673473544 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 6 |
Hb_000003_230 |
0.067467299 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002169_040 |
0.067994216 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_008841_020 |
0.0704578749 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002600_090 |
0.0716480393 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_006420_080 |
0.073465483 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_034380_020 |
0.0734838803 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 12 |
Hb_001828_150 |
0.0748481297 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000260_030 |
0.0754210923 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 14 |
Hb_000207_050 |
0.0767981646 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000827_050 |
0.0768231345 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_003768_030 |
0.0774374047 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_003697_040 |
0.0775267295 |
- |
- |
hypothetical protein B456_005G209600 [Gossypium raimondii] |
| 18 |
Hb_156279_020 |
0.0777065006 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 19 |
Hb_000288_030 |
0.0780544189 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 20 |
Hb_002799_070 |
0.0782394427 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |