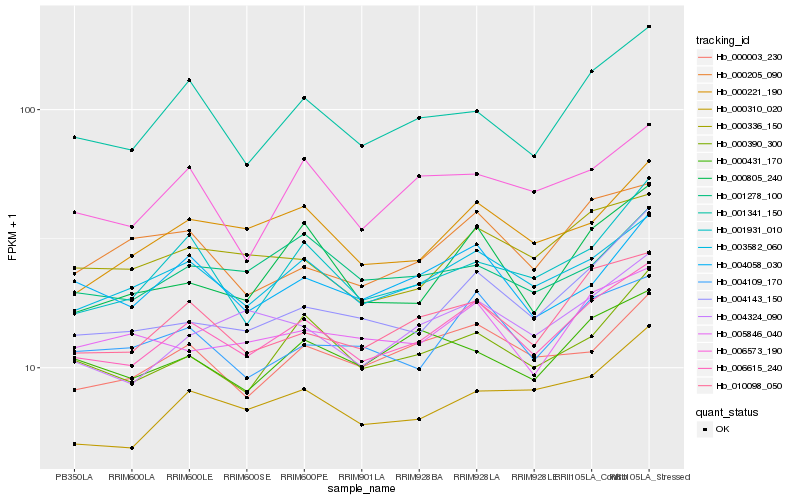
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006615_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 2 |
Hb_003582_060 |
0.0408222791 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_010098_050 |
0.0495702472 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 4 |
Hb_000390_300 |
0.0585286323 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 5 |
Hb_004109_170 |
0.0593506675 |
- |
- |
PREDICTED: probable ribokinase [Jatropha curcas] |
| 6 |
Hb_000003_230 |
0.0610507181 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001341_150 |
0.061129756 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 8 |
Hb_000336_150 |
0.0626819121 |
- |
- |
ribulose-5-phosphate-3-epimerase, putative [Ricinus communis] |
| 9 |
Hb_000221_190 |
0.0628653382 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 10 |
Hb_000310_020 |
0.0629776848 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 11 |
Hb_004058_030 |
0.0641188496 |
- |
- |
PREDICTED: stomatin-like protein 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_004143_150 |
0.0652921584 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 24-like isoform X6 [Jatropha curcas] |
| 13 |
Hb_005846_040 |
0.0657755912 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 4 [Populus euphratica] |
| 14 |
Hb_004324_090 |
0.0662576464 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000431_170 |
0.0682860791 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 16 |
Hb_006573_190 |
0.0686065066 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001278_100 |
0.0690490776 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 18 |
Hb_000805_240 |
0.069548135 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 19 |
Hb_001931_010 |
0.0701399635 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 20 |
Hb_000205_090 |
0.0702187569 |
- |
- |
hypothetical protein POPTR_0019s03570g [Populus trichocarpa] |