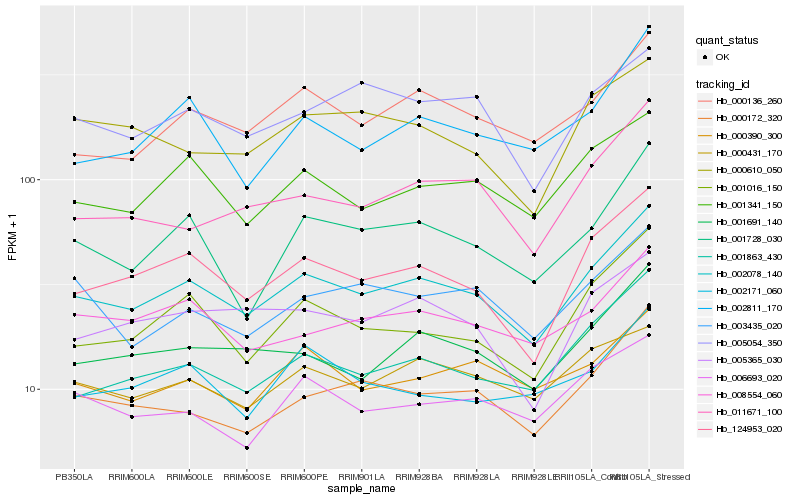
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_140 |
0.0 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_124953_020 |
0.0537349884 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 3 |
Hb_001691_140 |
0.0634056776 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 4 |
Hb_001341_150 |
0.0656162955 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 5 |
Hb_000136_260 |
0.0660194656 |
- |
- |
40S ribosomal protein S5A [Hevea brasiliensis] |
| 6 |
Hb_001016_150 |
0.066462921 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 7 |
Hb_001863_430 |
0.0677039745 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 8 |
Hb_000172_320 |
0.0681854856 |
- |
- |
PREDICTED: ribosome biogenesis protein TSR3 homolog [Jatropha curcas] |
| 9 |
Hb_001728_030 |
0.0716838717 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 10 |
Hb_008554_060 |
0.0725297179 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SPF27 homolog [Jatropha curcas] |
| 11 |
Hb_000431_170 |
0.0753208153 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 12 |
Hb_011671_100 |
0.0766646885 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 13 |
Hb_005365_030 |
0.0769824328 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 14 |
Hb_006693_020 |
0.077312405 |
- |
- |
PREDICTED: frataxin, mitochondrial [Gossypium raimondii] |
| 15 |
Hb_000390_300 |
0.0775503169 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 16 |
Hb_005054_350 |
0.0784546702 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P0 [Vitis vinifera] |
| 17 |
Hb_003435_020 |
0.0792852013 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_002811_170 |
0.0818590577 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 19 |
Hb_000610_050 |
0.0819084597 |
- |
- |
skp1, putative [Ricinus communis] |
| 20 |
Hb_002171_060 |
0.0822222676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |