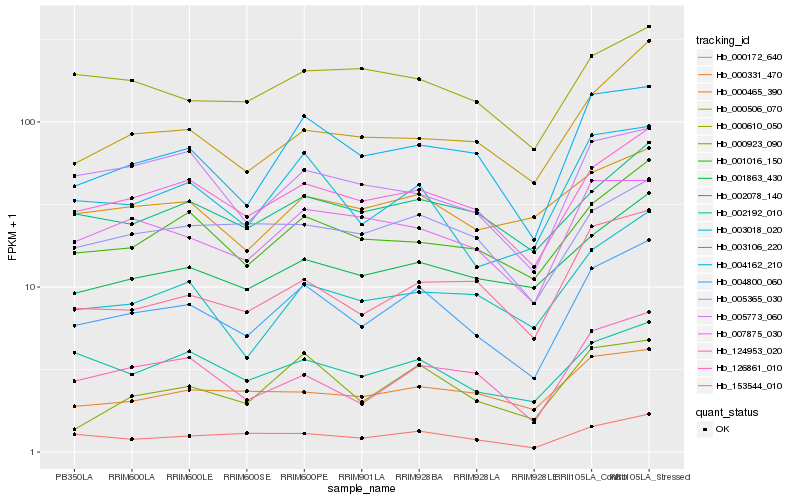
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 2 |
Hb_124953_020 |
0.0746611676 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 3 |
Hb_004162_210 |
0.0841131956 |
- |
- |
Cyclin-dependent kinases regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_003106_220 |
0.0885829382 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001016_150 |
0.0949467849 |
- |
- |
Rop1 [Hevea brasiliensis] |
| 6 |
Hb_000172_640 |
0.0951477736 |
- |
- |
hypothetical protein POPTR_0007s06250g [Populus trichocarpa] |
| 7 |
Hb_126861_010 |
0.1006205881 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 8 |
Hb_001863_430 |
0.1035480173 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 9 |
Hb_002078_140 |
0.104212363 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_007875_030 |
0.1045888436 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 11 |
Hb_153544_010 |
0.1062407345 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 12 |
Hb_002192_010 |
0.1063538065 |
- |
- |
- |
| 13 |
Hb_000506_070 |
0.1073467187 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 14 |
Hb_003018_020 |
0.109304151 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |
| 15 |
Hb_000923_090 |
0.1123169486 |
- |
- |
- |
| 16 |
Hb_005365_030 |
0.1154483181 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 17 |
Hb_000331_470 |
0.1174040123 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 18 |
Hb_000465_390 |
0.1184779212 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Jatropha curcas] |
| 19 |
Hb_000610_050 |
0.1192456694 |
- |
- |
skp1, putative [Ricinus communis] |
| 20 |
Hb_005773_060 |
0.120554845 |
- |
- |
defender against cell death, putative [Ricinus communis] |