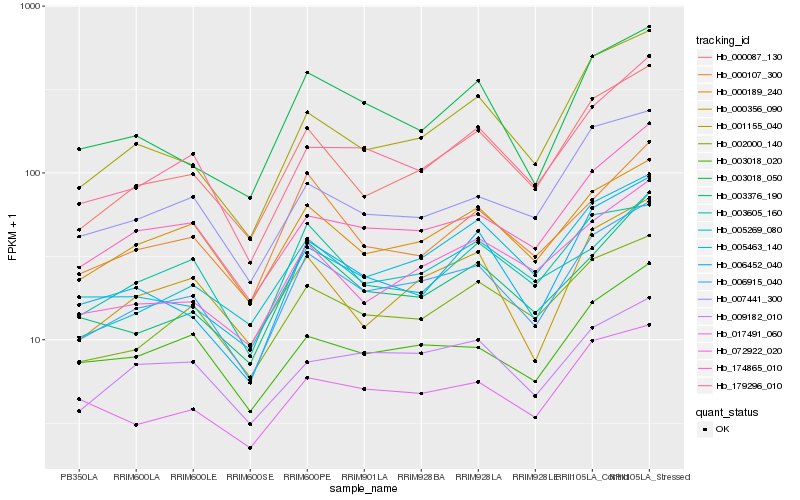
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006915_040 |
0.0 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 2 |
Hb_000087_130 |
0.0671451842 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000189_240 |
0.0730721261 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 4 |
Hb_002000_140 |
0.0763977405 |
- |
- |
PREDICTED: thioredoxin Y2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_179296_010 |
0.0800776434 |
- |
- |
hypothetical protein B456_006G094200 [Gossypium raimondii] |
| 6 |
Hb_174865_010 |
0.0825649076 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 7 |
Hb_003376_190 |
0.0855759861 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_001155_040 |
0.0866076109 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 9 |
Hb_072922_020 |
0.0872519243 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 10 |
Hb_000356_090 |
0.087704367 |
- |
- |
PREDICTED: uncharacterized protein At2g38710 [Jatropha curcas] |
| 11 |
Hb_003018_050 |
0.0881616153 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 12 |
Hb_017491_060 |
0.0898063736 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003018_020 |
0.0911155969 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |
| 14 |
Hb_007441_300 |
0.0920566364 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_005463_140 |
0.0930524823 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 16 |
Hb_006452_040 |
0.0932004069 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 17 |
Hb_000107_300 |
0.0938299209 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 [Jatropha curcas] |
| 18 |
Hb_005269_080 |
0.0954000774 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_009182_010 |
0.0954133679 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 20 |
Hb_003605_160 |
0.0954930166 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |