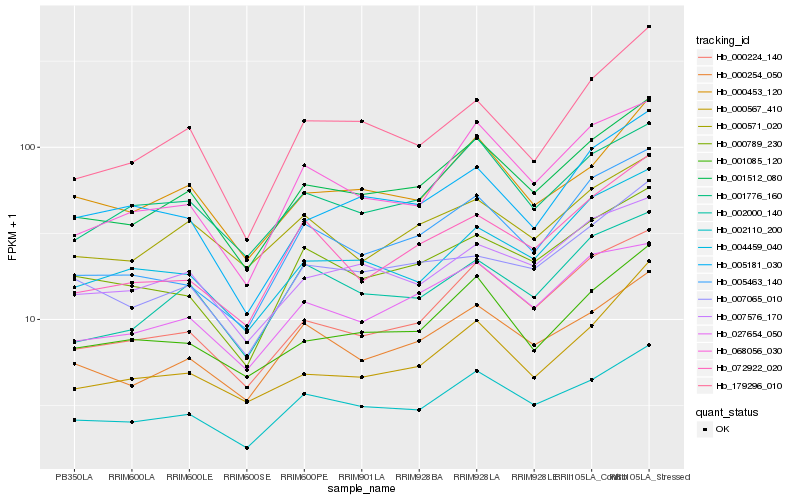
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001512_080 |
0.0 |
- |
- |
PREDICTED: proteasome subunit beta type-7-B [Jatropha curcas] |
| 2 |
Hb_002110_200 |
0.0558747265 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000224_140 |
0.0567481211 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_000254_050 |
0.0596144073 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 5 |
Hb_001085_120 |
0.065180666 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_027654_050 |
0.0695465104 |
- |
- |
PREDICTED: uncharacterized protein LOC105646592 [Jatropha curcas] |
| 7 |
Hb_000453_120 |
0.0695996258 |
- |
- |
PREDICTED: proteasome subunit alpha type-3 [Jatropha curcas] |
| 8 |
Hb_001776_160 |
0.0699460091 |
- |
- |
20S proteasome alpha subunit G1 [Theobroma cacao] |
| 9 |
Hb_007576_170 |
0.0732223327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_068056_030 |
0.0737327057 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 11 |
Hb_002000_140 |
0.0743417717 |
- |
- |
PREDICTED: thioredoxin Y2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005463_140 |
0.0753063843 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 13 |
Hb_004459_040 |
0.0753516582 |
- |
- |
PREDICTED: uncharacterized protein LOC105649649 [Jatropha curcas] |
| 14 |
Hb_000789_230 |
0.0769800097 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 15 |
Hb_007065_010 |
0.0774217414 |
- |
- |
hypothetical protein Csa_3G379730 [Cucumis sativus] |
| 16 |
Hb_179296_010 |
0.0831569446 |
- |
- |
hypothetical protein B456_006G094200 [Gossypium raimondii] |
| 17 |
Hb_000571_020 |
0.0834361714 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005181_030 |
0.0838204108 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 19 |
Hb_072922_020 |
0.0839411639 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 20 |
Hb_000567_410 |
0.0850001504 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |