| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
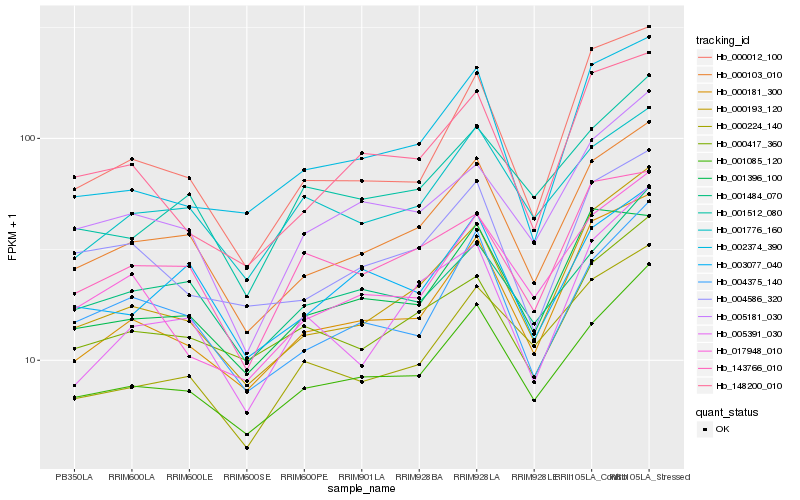
Hb_000103_010 |
0.0 |
- |
- |
20S proteasome beta subunit D2 [Hevea brasiliensis] |
| 2 |
Hb_000193_120 |
0.0533647907 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001085_120 |
0.0618131009 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 4 |
Hb_000181_300 |
0.0642938125 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004375_140 |
0.069838269 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003077_040 |
0.0762614825 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 7 |
Hb_005181_030 |
0.0791526603 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 8 |
Hb_000012_100 |
0.0813788848 |
- |
- |
UPF0587 protein C1orf123 homolog [Jatropha curcas] |
| 9 |
Hb_004586_320 |
0.0818079876 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 10 |
Hb_001396_100 |
0.0826507387 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001776_160 |
0.0843973399 |
- |
- |
20S proteasome alpha subunit G1 [Theobroma cacao] |
| 12 |
Hb_000417_360 |
0.085760938 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001512_080 |
0.086942177 |
- |
- |
PREDICTED: proteasome subunit beta type-7-B [Jatropha curcas] |
| 14 |
Hb_143766_010 |
0.0875257496 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 15 |
Hb_005391_030 |
0.0879587412 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 16 |
Hb_001484_070 |
0.0892902902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_148200_010 |
0.0895232542 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 18 |
Hb_000224_140 |
0.0897324159 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 19 |
Hb_002374_390 |
0.090245217 |
- |
- |
vacuolar ATP synthase proteolipid subunit 1, 2, 3, putative [Ricinus communis] |
| 20 |
Hb_017948_010 |
0.091758189 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |