| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
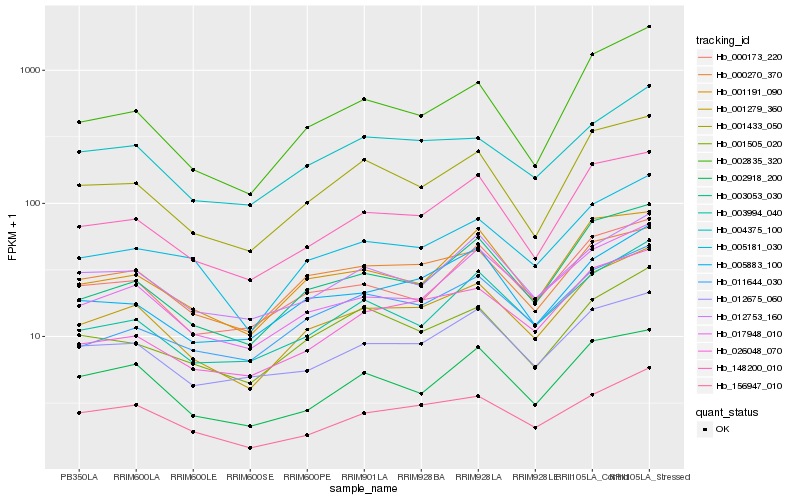
Hb_001279_360 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_148200_010 |
0.0647826745 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 3 |
Hb_017948_010 |
0.0666777862 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 4 |
Hb_003053_030 |
0.0681853257 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 5 |
Hb_005181_030 |
0.0683145185 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 6 |
Hb_156947_010 |
0.0733425395 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 7 |
Hb_001433_050 |
0.0737535942 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 8 |
Hb_001191_090 |
0.0791190859 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 9 |
Hb_000173_220 |
0.0791354109 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 10 |
Hb_004375_100 |
0.0792505435 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 11 |
Hb_005883_100 |
0.0824005032 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 12 |
Hb_003994_040 |
0.0824262538 |
- |
- |
- |
| 13 |
Hb_012675_060 |
0.0833046687 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 14 |
Hb_002835_320 |
0.0838486117 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 15 |
Hb_026048_070 |
0.0860068859 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 16 |
Hb_001505_020 |
0.0866889369 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 9 [Jatropha curcas] |
| 17 |
Hb_000270_370 |
0.0868405467 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 18 |
Hb_012753_160 |
0.0869966346 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_002918_200 |
0.0872435542 |
- |
- |
- |
| 20 |
Hb_011644_030 |
0.0875107159 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |