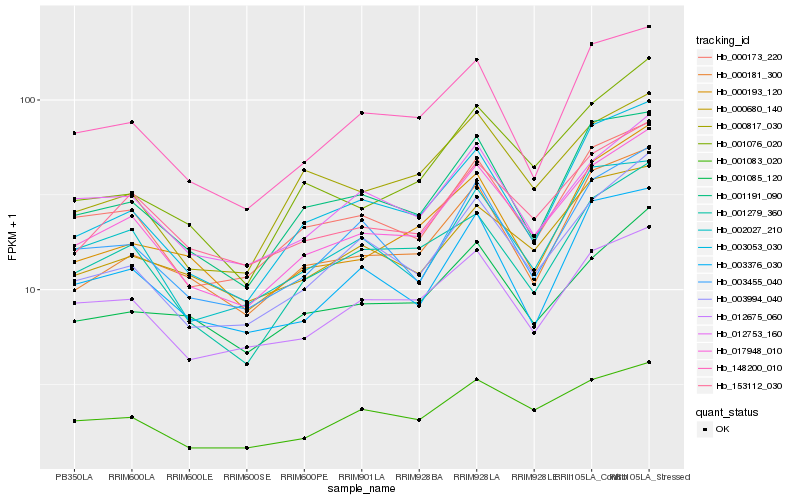
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017948_010 |
0.0 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 2 |
Hb_000173_220 |
0.0545823681 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 3 |
Hb_003994_040 |
0.0592331435 |
- |
- |
- |
| 4 |
Hb_153112_030 |
0.0593894607 |
- |
- |
PREDICTED: UPF0678 fatty acid-binding protein-like protein At1g79260 [Jatropha curcas] |
| 5 |
Hb_001279_360 |
0.0666777862 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001083_020 |
0.0683799914 |
- |
- |
- |
| 7 |
Hb_003053_030 |
0.0685143823 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 8 |
Hb_001191_090 |
0.0715591503 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 9 |
Hb_012753_160 |
0.0721670796 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 10 |
Hb_012675_060 |
0.0727876661 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 11 |
Hb_000181_300 |
0.0736472186 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 12 |
Hb_148200_010 |
0.0747775936 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 13 |
Hb_003455_040 |
0.0753214271 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 14 |
Hb_002027_210 |
0.0771726349 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 15 |
Hb_001085_120 |
0.0777818761 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_000680_140 |
0.080202349 |
- |
- |
PREDICTED: uncharacterized protein LOC105631472 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000817_030 |
0.0805863661 |
- |
- |
mak, putative [Ricinus communis] |
| 18 |
Hb_001076_020 |
0.080987965 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 19 |
Hb_000193_120 |
0.0829716381 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 20 |
Hb_003376_030 |
0.0848223288 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |