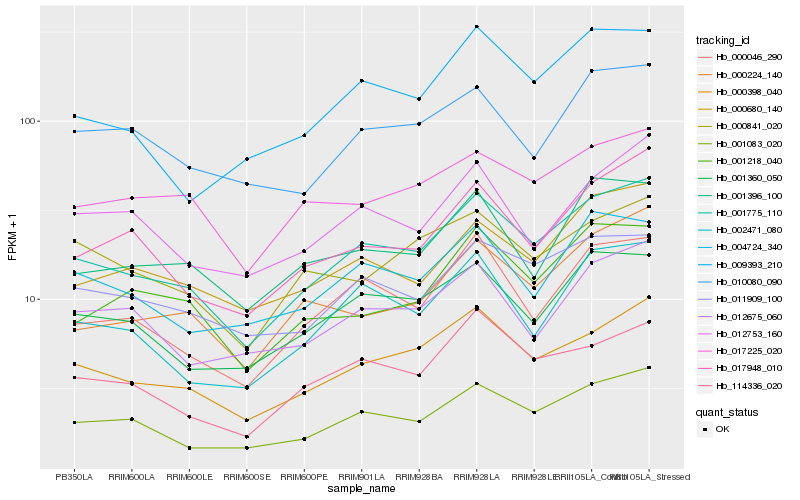
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_110 |
0.0 |
- |
- |
PREDICTED: probable ribosome biogenesis protein RLP24 [Jatropha curcas] |
| 2 |
Hb_000398_040 |
0.0578737747 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 3 |
Hb_001083_020 |
0.0584318296 |
- |
- |
- |
| 4 |
Hb_000046_290 |
0.0764761522 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 5 |
Hb_001360_050 |
0.08107723 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011909_100 |
0.082752095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_114336_020 |
0.0836700957 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 8 |
Hb_017948_010 |
0.0856234493 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 9 |
Hb_002471_080 |
0.085870171 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 10 |
Hb_000680_140 |
0.0878395134 |
- |
- |
PREDICTED: uncharacterized protein LOC105631472 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009393_210 |
0.0894607486 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 12 |
Hb_004724_340 |
0.0911736787 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 13 |
Hb_012675_060 |
0.094398672 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 14 |
Hb_010080_090 |
0.0945335277 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 [Jatropha curcas] |
| 15 |
Hb_001218_040 |
0.0947886608 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 16 |
Hb_000224_140 |
0.0950343506 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 17 |
Hb_017225_020 |
0.0952850193 |
- |
- |
PREDICTED: protein BRICK 1 [Gossypium raimondii] |
| 18 |
Hb_001396_100 |
0.0953291481 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_012753_160 |
0.0967386459 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 20 |
Hb_000841_020 |
0.096776601 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |