| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
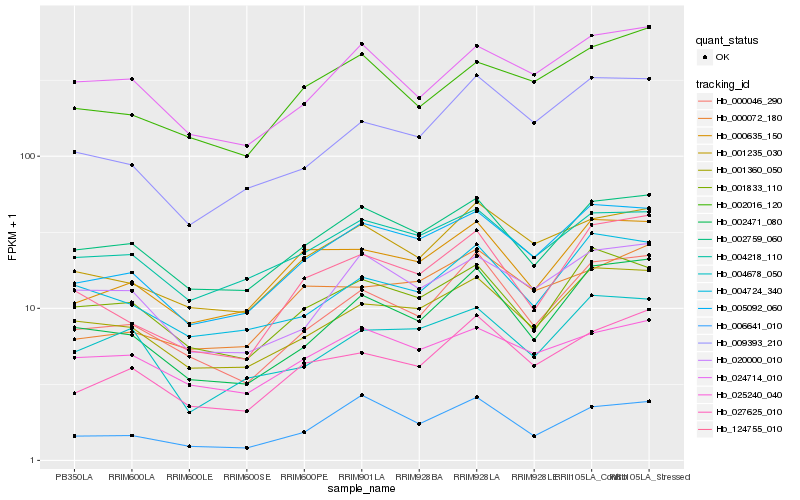
Hb_005092_060 |
0.0 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 2 |
Hb_001360_050 |
0.0694516771 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001235_030 |
0.0714775579 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 4 |
Hb_002759_060 |
0.0716223595 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 5 |
Hb_000635_150 |
0.0733833681 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 6 |
Hb_004218_110 |
0.0750672254 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 7 |
Hb_001833_110 |
0.0849311902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_027625_010 |
0.0910024324 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 9 |
Hb_000046_290 |
0.0917086525 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 10 |
Hb_020000_010 |
0.0922666316 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 11 |
Hb_009393_210 |
0.0929585051 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 12 |
Hb_000072_180 |
0.093470052 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 13 |
Hb_002471_080 |
0.0936949595 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 14 |
Hb_006641_010 |
0.0953217109 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 15 |
Hb_002016_120 |
0.0965181598 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940 [Jatropha curcas] |
| 16 |
Hb_124755_010 |
0.0970552491 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 17 |
Hb_004678_050 |
0.0975230619 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase [Jatropha curcas] |
| 18 |
Hb_024714_010 |
0.0984839608 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_025240_040 |
0.0986622191 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |
| 20 |
Hb_004724_340 |
0.0989964378 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |