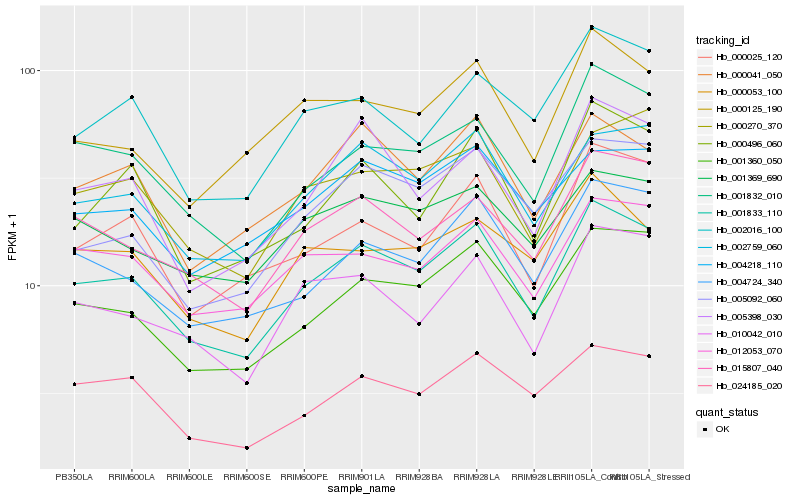
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001833_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002759_060 |
0.0614455109 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 3 |
Hb_001360_050 |
0.0630067793 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015807_040 |
0.069772802 |
- |
- |
PREDICTED: polycomb group protein FERTILIZATION-INDEPENDENT ENDOSPERM [Jatropha curcas] |
| 5 |
Hb_001832_010 |
0.0704706913 |
- |
- |
PREDICTED: uncharacterized protein LOC105637890 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004724_340 |
0.0730666277 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 7 |
Hb_000041_050 |
0.0762114853 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 8 |
Hb_012053_070 |
0.0771413427 |
- |
- |
PREDICTED: uncharacterized protein LOC105637344 [Jatropha curcas] |
| 9 |
Hb_024185_020 |
0.0787000678 |
- |
- |
- |
| 10 |
Hb_010042_010 |
0.0787618703 |
- |
- |
Urease accessory protein ureD, putative [Ricinus communis] |
| 11 |
Hb_001369_690 |
0.0805937077 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 12 |
Hb_005398_030 |
0.080968357 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase 1-like [Jatropha curcas] |
| 13 |
Hb_000025_120 |
0.0819427792 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004218_110 |
0.0822959687 |
- |
- |
hypothetical protein PRUPE_ppa006329mg [Prunus persica] |
| 15 |
Hb_000496_060 |
0.0840825778 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 16 |
Hb_000270_370 |
0.0841740368 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 17 |
Hb_000125_190 |
0.0842088119 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002016_100 |
0.08468555 |
- |
- |
Small nuclear ribonucleoprotein SM D3, putative [Ricinus communis] |
| 19 |
Hb_005092_060 |
0.0849311902 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 20 |
Hb_000053_100 |
0.0850571317 |
- |
- |
PREDICTED: uncharacterized protein LOC105642071 [Jatropha curcas] |