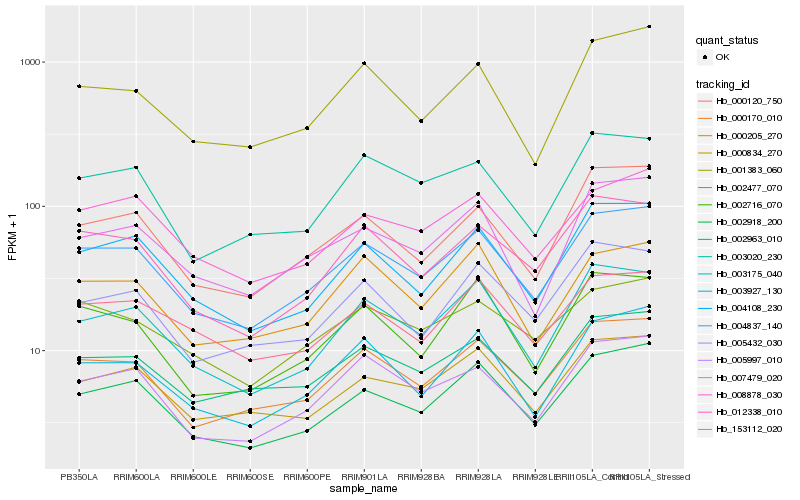
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_140 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 2 |
Hb_000170_010 |
0.0437533822 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002918_200 |
0.0512693905 |
- |
- |
- |
| 4 |
Hb_002963_010 |
0.0574805421 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004108_230 |
0.0576683205 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 6 |
Hb_005997_010 |
0.0593614734 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 7 |
Hb_012338_010 |
0.0617437695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000120_750 |
0.0638288819 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_003175_040 |
0.0642434999 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000834_270 |
0.0643582866 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 11 |
Hb_008878_030 |
0.0661615793 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Musa acuminata subsp. malaccensis] |
| 12 |
Hb_003927_130 |
0.0681400145 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 13 |
Hb_001383_060 |
0.0693424655 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 14 |
Hb_007479_020 |
0.0698959827 |
- |
- |
PREDICTED: CMP-sialic acid transporter 5 [Jatropha curcas] |
| 15 |
Hb_003020_230 |
0.0722348791 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 16 |
Hb_002477_070 |
0.072423831 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 17 |
Hb_002716_070 |
0.0755313063 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 18 |
Hb_000205_270 |
0.0760155248 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 19 |
Hb_153112_020 |
0.0763940053 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 22b-like [Jatropha curcas] |
| 20 |
Hb_005432_030 |
0.0768740629 |
- |
- |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Gossypium raimondii] |