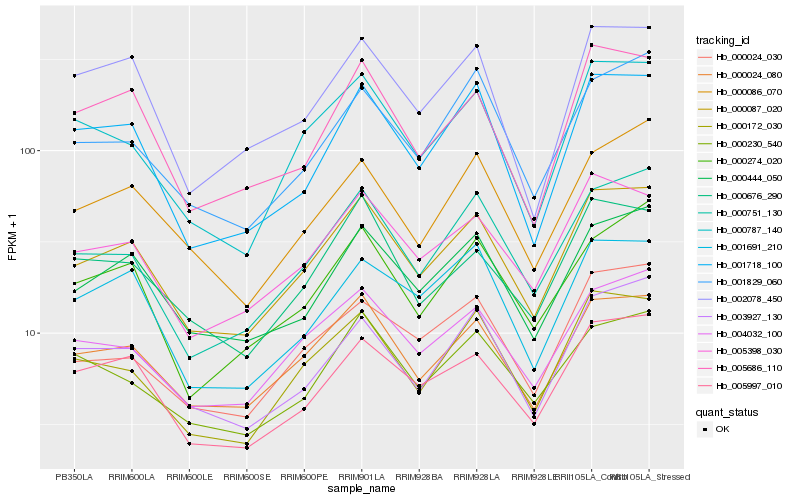
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_020 |
0.0 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000024_080 |
0.04323019 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000444_050 |
0.0574524582 |
transcription factor |
TF Family: HMG |
PREDICTED: uncharacterized protein LOC105648212 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005398_030 |
0.057733736 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase 1-like [Jatropha curcas] |
| 5 |
Hb_004032_100 |
0.0604822052 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like [Jatropha curcas] |
| 6 |
Hb_005997_010 |
0.0637571128 |
- |
- |
Dihydroorotate dehydrogenase (quinone) [Morus notabilis] |
| 7 |
Hb_000751_130 |
0.0639548922 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 8 |
Hb_000172_030 |
0.0681664419 |
- |
- |
PREDICTED: protein SCO1 homolog 2, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000787_140 |
0.0720761826 |
- |
- |
Trafficking protein particle complex subunit, putative [Ricinus communis] |
| 10 |
Hb_001718_100 |
0.0760479243 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 11 |
Hb_000676_290 |
0.0765806238 |
- |
- |
PREDICTED: glutaredoxin-C3 [Jatropha curcas] |
| 12 |
Hb_005686_110 |
0.0768300474 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 13 |
Hb_000274_020 |
0.0787423273 |
- |
- |
PREDICTED: uncharacterized protein LOC105630805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003927_130 |
0.0788168192 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 15 |
Hb_000024_030 |
0.0799418009 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 5-like [Populus euphratica] |
| 16 |
Hb_000086_070 |
0.0801307605 |
- |
- |
Peptidase S24/S26A/S26B/S26C family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_001691_210 |
0.0805693122 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 18 |
Hb_001829_060 |
0.0820882912 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 19 |
Hb_000230_540 |
0.0824382577 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 3, mitochondrial [Sesamum indicum] |
| 20 |
Hb_002078_450 |
0.0834836241 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |