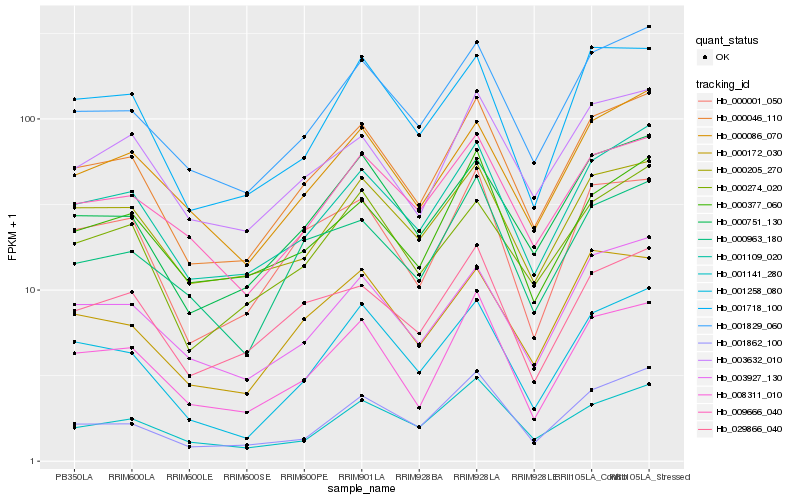
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_110 |
0.0 |
- |
- |
PREDICTED: 54S ribosomal protein L39, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001109_020 |
0.0502126166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001829_060 |
0.0548824685 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 4 |
Hb_000751_130 |
0.0649053418 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit tim16 [Jatropha curcas] |
| 5 |
Hb_008311_010 |
0.0724547397 |
- |
- |
hypothetical protein CARUB_v10003781mg [Capsella rubella] |
| 6 |
Hb_003632_010 |
0.0734505277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003927_130 |
0.0754700256 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 8 |
Hb_000001_050 |
0.0758194148 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001258_080 |
0.0764246394 |
- |
- |
PREDICTED: uncharacterized protein LOC105639921 [Jatropha curcas] |
| 10 |
Hb_000205_270 |
0.0777777405 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000963_180 |
0.0788647429 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 12 |
Hb_000274_020 |
0.0795446745 |
- |
- |
PREDICTED: uncharacterized protein LOC105630805 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000086_070 |
0.0796108654 |
- |
- |
Peptidase S24/S26A/S26B/S26C family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_029866_040 |
0.0809667356 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 15 |
Hb_009666_040 |
0.0810784998 |
- |
- |
glutathione transferase, putative [Ricinus communis] |
| 16 |
Hb_000377_060 |
0.0812668234 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 17 |
Hb_000172_030 |
0.0829595176 |
- |
- |
PREDICTED: protein SCO1 homolog 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001141_280 |
0.0831312189 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 19 |
Hb_001862_100 |
0.0831886406 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001718_100 |
0.0836412113 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |